A Robust One-Step Recombineering System for Enterohemorrhagic Escherichia coli
- PMID: 36144292
- PMCID: PMC9504302
- DOI: 10.3390/microorganisms10091689
A Robust One-Step Recombineering System for Enterohemorrhagic Escherichia coli
Abstract
Enterohemorrhagic Escherichia coli (EHEC) can cause severe diarrheic in humans. To improve therapy options, a better understanding of EHEC pathogenicity is essential. The genetic manipulation of EHEC with classical one-step methods, such as the transient overexpression of the phage lambda (λ) Red functions, is not very efficient. Here, we provide a robust and reliable method for increasing recombineering efficiency in EHEC based on the transient coexpression of recX together with gam, beta, and exo. We demonstrate that the genetic manipulation is 3-4 times more efficient in EHEC O157:H7 EDL933 Δstx1/2 with our method when compared to the overexpression of the λ Red functions alone. Both recombineering systems demonstrated similar efficiencies in Escherichia coli K-12 MG1655. Coexpression of recX did not enhance the Gam-mediated inhibition of sparfloxacin-mediated SOS response. Therefore, the additional inhibition of the RecFOR pathway rather than a stronger inhibition of the RecBCD pathway of SOS response induction might have resulted in the increased recombineering efficiency by indirectly blocking phage induction. Even though additional experiments are required to unravel the precise mechanistic details of the improved recombineering efficiency, we recommend the use of our method for the robust genetic manipulation of EHEC and other prophage-carrying E. coli isolates.
Keywords: SOS response; enterohemorrhagic Escherichia coli; recombineering.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
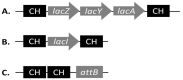
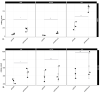
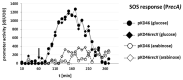
References
-
- Michino H., Araki K., Minami S., Takaya S., Sakai N., Miyazaki M., Ono A., Yanagawa H. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 1999;150:787–796. doi: 10.1093/oxfordjournals.aje.a010082. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

