Phylogeny of Regulators of G-Protein Signaling Genes in Leptographium qinlingensis and Expression Levels of Three RGSs in Response to Different Terpenoids
- PMID: 36144299
- PMCID: PMC9506272
- DOI: 10.3390/microorganisms10091698
Phylogeny of Regulators of G-Protein Signaling Genes in Leptographium qinlingensis and Expression Levels of Three RGSs in Response to Different Terpenoids
Abstract
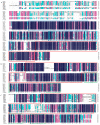
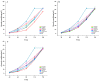
Leptographium qinlingensis is a bark beetle-vectored pine pathogen in the Chinese white pine beetle (Dendroctonus armandi) epidemic in Northwest China. L. qinlingensis colonizes pines despite the trees' massive oleoresin terpenoid defenses. Regulators of G-protein signaling (RGS) proteins modulate heterotrimeric G-protein signaling negatively and play multiple roles in the growth, asexual development, and pathogenicity of fungi. In this study, we have identified three L. qinlingensis RGS genes, and the phylogenetic analysis shows the highest homology with the regulators of G-protein signaling proteins sequence from Ophiostoma piceae and Grosmannia clavigera. The expression profiles of three RGSs in the mycelium of L. qinlingensis treated with six different terpenoids were detected, as well as their growth rates. Under six terpenoid treatments, the growth and reproduction in L. qinlingensis were significantly inhibited, and the growth inflection day was delayed from 8 days to 12-13 days. By analyzing the expression level of three RGS genes of L. qinlingensis with different treatments, results indicate that LqFlbA plays a crucial role in controlling fungal growth, and both LqRax1 and LqRgsA are involved in overcoming the host chemical resistances and successful colonization.
Keywords: Leptographium qinlingensis; expression; host resistances; regulators of G-protein signaling; terpene tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Phylogeny of Leptographium qinlingensis cytochrome P450 genes and transcription levels of six CYPs in response to different nutrition media or terpenoids.Arch Microbiol. 2021 Dec 11;204(1):16. doi: 10.1007/s00203-021-02616-9. Arch Microbiol. 2021. PMID: 34894279
-
Molecular Mechanism of Overcoming Host Resistance by the Target of Rapamycin Gene in Leptographium qinlingensis.Microorganisms. 2022 Feb 24;10(3):503. doi: 10.3390/microorganisms10030503. Microorganisms. 2022. PMID: 35336079 Free PMC article.
-
The CYP51F1 Gene of Leptographium qinlingensis: Sequence Characteristic, Phylogeny and Transcript Levels.Int J Mol Sci. 2015 May 26;16(6):12014-34. doi: 10.3390/ijms160612014. Int J Mol Sci. 2015. PMID: 26016505 Free PMC article.
-
Mountain Pine Beetle Epidemic: An Interplay of Terpenoids in Host Defense and Insect Pheromones.Annu Rev Plant Biol. 2022 May 20;73:475-494. doi: 10.1146/annurev-arplant-070921-103617. Epub 2022 Feb 7. Annu Rev Plant Biol. 2022. PMID: 35130442 Review.
-
Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans.J Microbiol. 2006 Apr;44(2):145-54. J Microbiol. 2006. PMID: 16728950 Review.
Cited by
-
Establishment of RNA Interference Genetic Transformation System and Functional Analysis of FlbA Gene in Leptographium qinlingensis.Int J Mol Sci. 2023 Aug 21;24(16):13009. doi: 10.3390/ijms241613009. Int J Mol Sci. 2023. PMID: 37629189 Free PMC article.
References
-
- Hu X., Yu J., Wang C., Chen H. Cellulolytic bacteria associated with the gut of Dendroctonus armandi larvae (Coleoptera: Curculionidae: Scolytinae) Forests. 2014;5:455–465. doi: 10.3390/f5030455. - DOI
-
- Chen H., Tang M., Zhu C., Hu J. The enzymes in the secretions of Dendroctonus armandi (Scolytidae) and their symbiotic fungus of Leptographium qinlingensis. Sci. Silvae Sin. 2004;40:123–126. (In Chinese)
-
- Tang M., Chen H. Effect of symbiotic fungi of Dendroctonus armandi on host trees. Sci. Silvae Sin. 1999;35:63–66. (In Chinese)
Grants and funding
LinkOut - more resources
Full Text Sources

