Morganella Phage Mecenats66 Utilizes an Evolutionarily Distinct Subtype of Headful Genome Packaging with a Preferred Packaging Initiation Site
- PMID: 36144401
- PMCID: PMC9503643
- DOI: 10.3390/microorganisms10091799
Morganella Phage Mecenats66 Utilizes an Evolutionarily Distinct Subtype of Headful Genome Packaging with a Preferred Packaging Initiation Site
Abstract
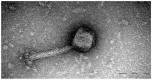
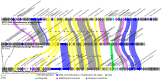
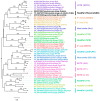
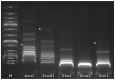
Both recognized species from the genus Morganella (M. morganii and M. psychrotolerans) are Gram-negative facultative anaerobic rod-shaped bacteria that have been documented as sometimes being implicated in human disease. Complete genomes of seven Morganella-infecting phages are publicly available today. Here, we report on the genomic characterization of an insect associated Morganella sp. phage, which we named Mecenats66, isolated from dead worker honeybees. Phage Mecenats66 was propagated, purified, and subjected to whole-genome sequencing with subsequent complete genome annotation. After the genome de novo assembly, it was noted that Mecenats66 might employ a headful packaging with a preferred packaging initiation site, although its terminase amino acid sequence did not fall within any of the currently recognized headful packaging subtype employing phage (that had their packaging strategy experimentally verified) with clusters on a terminase sequence phylogenetic tree. The in silico predicted packaging strategy was verified experimentally, validating the packaging initiation site and suggesting that Mecenats66 represents an evolutionarily distinct headful genome packaging with a preferred packaging initiation site strategy subtype. These findings can possibly be attributed to several of the phages already found within the public biological sequence repositories and could aid newly isolated phage packaging strategy predictions in the future.
Keywords: Morganella; bacteriophage; complete genome; genome termini; headful packaging; packaging initiation (pac) site; terminase.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

