Genomic Adaptations of Saccharomyces Genus to Wine Niche
- PMID: 36144411
- PMCID: PMC9500811
- DOI: 10.3390/microorganisms10091811
Genomic Adaptations of Saccharomyces Genus to Wine Niche
Abstract
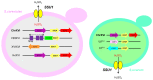
Wine yeast have been exposed to harsh conditions for millennia, which have led to adaptive evolutionary strategies. Thus, wine yeasts from Saccharomyces genus are considered an interesting and highly valuable model to study human-drive domestication processes. The rise of whole-genome sequencing technologies together with new long reads platforms has provided new understanding about the population structure and the evolution of wine yeasts. Population genomics studies have indicated domestication fingerprints in wine yeast, including nucleotide variations, chromosomal rearrangements, horizontal gene transfer or hybridization, among others. These genetic changes contribute to genetically and phenotypically distinct strains. This review will summarize and discuss recent research on evolutionary trajectories of wine yeasts, highlighting the domestication hallmarks identified in this group of yeast.
Keywords: chromosomal rearrangements; copy number variation; domestication; horizontal gene transfer; hybridization; wine yeast.
Conflict of interest statement
The author declares no conflict of interest.
Figures

Similar articles
-
Diversity and adaptive evolution of Saccharomyces wine yeast: a review.FEMS Yeast Res. 2015 Nov;15(7):fov067. doi: 10.1093/femsyr/fov067. Epub 2015 Jul 22. FEMS Yeast Res. 2015. PMID: 26205244 Free PMC article. Review.
-
Distinct Domestication Trajectories in Top-Fermenting Beer Yeasts and Wine Yeasts.Curr Biol. 2016 Oct 24;26(20):2750-2761. doi: 10.1016/j.cub.2016.08.040. Epub 2016 Oct 6. Curr Biol. 2016. PMID: 27720622
-
Genomics and Biochemistry of Saccharomyces cerevisiae Wine Yeast Strains.Biochemistry (Mosc). 2016 Dec;81(13):1650-1668. doi: 10.1134/S0006297916130046. Biochemistry (Mosc). 2016. PMID: 28260488 Review.
-
Genomic signatures of adaptation to wine biological ageing conditions in biofilm-forming flor yeasts.Mol Ecol. 2017 Apr;26(7):2150-2166. doi: 10.1111/mec.14053. Epub 2017 Mar 13. Mol Ecol. 2017. PMID: 28192619
-
A population genomics insight into the Mediterranean origins of wine yeast domestication.Mol Ecol. 2015 Nov;24(21):5412-27. doi: 10.1111/mec.13341. Epub 2015 Sep 3. Mol Ecol. 2015. PMID: 26248006
Cited by
-
Predictability of the community-function landscape in wine yeast ecosystems.Mol Syst Biol. 2023 Sep 12;19(9):e11613. doi: 10.15252/msb.202311613. Epub 2023 Aug 7. Mol Syst Biol. 2023. PMID: 37548146 Free PMC article.
-
Fermentative factors shape transcriptional response of Lachancea thermotolerans and wine acidification.NPJ Sci Food. 2025 Jun 6;9(1):97. doi: 10.1038/s41538-025-00467-y. NPJ Sci Food. 2025. PMID: 40481005 Free PMC article.
-
Subpopulation-specific gene expression in Lachancea thermotolerans uncovers distinct metabolic adaptations to wine fermentation.Curr Res Food Sci. 2024 Dec 11;10:100954. doi: 10.1016/j.crfs.2024.100954. eCollection 2025. Curr Res Food Sci. 2024. PMID: 39760014 Free PMC article.
-
Ecology and Evolutionary Biology as Frameworks to Study Wine Fermentations.Microb Biotechnol. 2025 Mar;18(3):e70078. doi: 10.1111/1751-7915.70078. Microb Biotechnol. 2025. PMID: 40136006 Free PMC article. Review.
-
Renewing Lost Genetic Variability with a Classical Yeast Genetics Approach.J Fungi (Basel). 2023 Feb 16;9(2):264. doi: 10.3390/jof9020264. J Fungi (Basel). 2023. PMID: 36836378 Free PMC article.
References
-
- Legras J., Galeote V., Bigey F., Camarasa C., Marsit S., Nidelet T., Sanchez I., Couloux A., Guy J., Franco-duarte R., et al. Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 2018;35:1712–1727. doi: 10.1093/molbev/msy066. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

