Extracellular Vesicle Subproteome Differences among Filifactor alocis Clinical Isolates
- PMID: 36144428
- PMCID: PMC9503520
- DOI: 10.3390/microorganisms10091826
Extracellular Vesicle Subproteome Differences among Filifactor alocis Clinical Isolates
Abstract
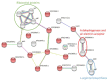
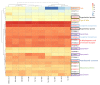
Filifactor alocis is a Gram-positive asaccharolytic, obligate anaerobic rod of the Firmicutes phylum, which has recently been implicated in oral infections. Extracellular vesicles (EVs) are crucial conveyors of microbial virulence in bacteria and archaea. Previously, in highly purified EVs from the F. alocis reference strain ATCC 35896 (CCUG 47790), 28 proteins were identified. The present study aimed to use label-free quantification proteomics in order to chart these EV proteins, in the reference strain, and in nine less-well-characterized clinical F. alocis isolates. In total, 25 of the EV proteins were identified and 24 were quantified. Sixteen of those were differentially expressed between the ten strains and the novel FtxA RTX toxin and one lipoprotein were among them. Consistent expression was observed among ribosomal proteins and proteins involved in L-arginine biosynthesis and type IV pilin, demonstrating a degree of EV protein expression preservation among strains. In terms of protein-protein interaction analysis, 21 functional associations were revealed between 19 EV proteins. Interestingly, FtxA did not display predicted interactions with any other EV protein. In conclusion, the present study charted 25 EV proteins in ten F. alocis strains. While most EV proteins were consistently identified among the strains, several of them were also differentially expressed, which justifies that there may be potential variations in the virulence potential among EVs of different F. alocis strains.
Keywords: Filifactor alocis; extracellular vesicles (EVs); oral infections; predicted EV subproteome; proteomics.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
Proteomic Characterization of the Oral Pathogen Filifactor alocis Reveals Key Inter-Protein Interactions of Its RTX Toxin: FtxA.Pathogens. 2022 May 17;11(5):590. doi: 10.3390/pathogens11050590. Pathogens. 2022. PMID: 35631111 Free PMC article.
-
Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA.Toxins (Basel). 2020 Oct 30;12(11):687. doi: 10.3390/toxins12110687. Toxins (Basel). 2020. PMID: 33143036 Free PMC article.
-
Characterization and immunostimulatory activity of extracellular vesicles from Filifactor alocis.Mol Oral Microbiol. 2020 Jan;35(1):1-9. doi: 10.1111/omi.12272. Epub 2019 Dec 3. Mol Oral Microbiol. 2020. PMID: 31675472
-
Filifactor alocis: The Newly Discovered Kid on the Block with Special Talents.J Dent Res. 2014 Aug;93(8):725-32. doi: 10.1177/0022034514538283. Epub 2014 Jun 4. J Dent Res. 2014. PMID: 24898946 Free PMC article. Review.
-
Aggregatibacter actinomycetemcomitans and Filifactor alocis: Two exotoxin-producing oral pathogens.Front Oral Health. 2022 Aug 15;3:981343. doi: 10.3389/froh.2022.981343. eCollection 2022. Front Oral Health. 2022. PMID: 36046121 Free PMC article. Review.
Cited by
-
Identification of Filifactor Alocis in Periodontal Biofilms using Polymerase Chain Reaction Technique: A Cross-Sectional Study.J Pharm Bioallied Sci. 2024 Dec;16(Suppl 5):S4381-S4386. doi: 10.4103/jpbs.jpbs_625_24. Epub 2025 Jan 30. J Pharm Bioallied Sci. 2024. PMID: 40061744 Free PMC article.
-
Whole Genome Sequencing and Phenotypic Analysis of Antibiotic Resistance in Filifactor alocis Isolates.Antibiotics (Basel). 2023 Jun 15;12(6):1059. doi: 10.3390/antibiotics12061059. Antibiotics (Basel). 2023. PMID: 37370380 Free PMC article.
-
Oral Microorganisms and Biofilms: New Insights to Defeat the Main Etiologic Factor of Oral Diseases.Microorganisms. 2022 Dec 6;10(12):2413. doi: 10.3390/microorganisms10122413. Microorganisms. 2022. PMID: 36557666 Free PMC article.
References
-
- Nadeem A., Oscarsson J., Wai S.N. Delivery of Virulence Factors by Bacterial Membrane Vesicles to Mammalian Host Cells. In: Kaparakis-Liaskos M., Kufer T., editors. Bacterial Membrane Vesicles. Springer; Cham, Switzerland: 2020. pp. 131–158. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

