Genome-Based Exploration of Rhodococcus Species for Plastic-Degrading Genetic Determinants Using Bioinformatic Analysis
- PMID: 36144448
- PMCID: PMC9506104
- DOI: 10.3390/microorganisms10091846
Genome-Based Exploration of Rhodococcus Species for Plastic-Degrading Genetic Determinants Using Bioinformatic Analysis
Abstract
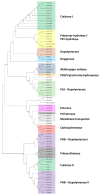
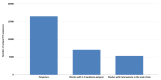
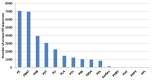
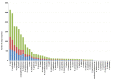
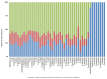
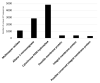
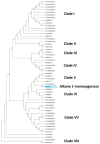
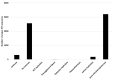
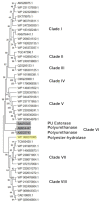
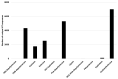
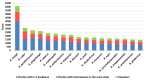
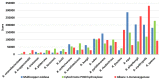
Plastic polymer waste management is an increasingly prevalent issue. In this paper, Rhodococcus genomes were explored to predict new plastic-degrading enzymes based on recently discovered biodegrading enzymes for diverse plastic polymers. Bioinformatics prediction analyses were conducted using 124 gene products deriving from diverse microorganisms retrieved from databases, literature data, omic-approaches, and functional analyses. The whole results showed the plastic-degrading potential of Rhodococcus genus. Among the species with high plastic-degrading potential, R. erythropolis, R. equi, R. opacus, R. qingshengii, R. fascians, and R. rhodochrous appeared to be the most promising for possible plastic removal. A high number of genetic determinants related to polyester biodegradation were obtained from different Rhodococcus species. However, score calculation demonstrated that Rhodococcus species (especially R. pyridinivorans, R. qingshengii, and R. hoagii) likely possess PE-degrading enzymes. The results identified diverse oxidative systems, including multicopper oxidases, alkane monooxygenases, cytochrome P450 hydroxylases, para-nitrobenzylesterase, and carboxylesterase, and they could be promising reference sequences for the biodegradation of plastics with C-C backbone, plastics with heteroatoms in the main chain, and polyesters, respectively. Notably, the results of this study could be further exploited for biotechnological applications in biodegradative processes using diverse Rhodococcus strains and through catalytic reactions.
Keywords: PET-hydrolase; Rhodococcus genus; depolymerase; esterase; genome analyses; hydroxylase/monooxygenase; oxidase; plastic; plastic-degrading enzymes; polymer biodegradation.
Conflict of interest statement
All other authors declare no conflict of interest.
Figures


Similar articles
-
Polyethylene Biodegradation by an Artificial Bacterial Consortium: Rhodococcus as a Competitive Plastisphere Species.Microbes Environ. 2024;39(3):ME24031. doi: 10.1264/jsme2.ME24031. Microbes Environ. 2024. PMID: 39085141 Free PMC article.
-
Insights into the biodegradation of polycaprolactone through genomic analysis of two plastic-degrading Rhodococcus bacteria.Front Microbiol. 2024 Jan 3;14:1284956. doi: 10.3389/fmicb.2023.1284956. eCollection 2023. Front Microbiol. 2024. PMID: 38235436 Free PMC article.
-
Transcriptomic analysis of Rhodococcus opacus R7 grown on polyethylene by RNA-seq.Sci Rep. 2021 Oct 29;11(1):21311. doi: 10.1038/s41598-021-00525-x. Sci Rep. 2021. PMID: 34716360 Free PMC article.
-
Can whole genome analysis refine the taxonomy of the genus Rhodococcus?FEMS Microbiol Rev. 2004 Jun;28(3):377-403. doi: 10.1016/j.femsre.2004.01.001. FEMS Microbiol Rev. 2004. PMID: 15449609 Review.
-
Genome analysis and -omics approaches provide new insights into the biodegradation potential of Rhodococcus.Appl Microbiol Biotechnol. 2019 Feb;103(3):1069-1080. doi: 10.1007/s00253-018-9539-7. Epub 2018 Dec 15. Appl Microbiol Biotechnol. 2019. PMID: 30554387 Review.
Cited by
-
Insights into polyethylene biodegradative fingerprint of Pseudomonas citronellolis E5 and Rhodococcus erythropolis D4 by phenotypic and genome-based comparative analyses.Front Bioeng Biotechnol. 2024 Dec 12;12:1472309. doi: 10.3389/fbioe.2024.1472309. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39726982 Free PMC article.
-
Insight on recently discovered PET polyester-degrading enzymes, thermostability and activity analyses.3 Biotech. 2024 Jan;14(1):31. doi: 10.1007/s13205-023-03882-8. Epub 2024 Jan 2. 3 Biotech. 2024. PMID: 38178895 Free PMC article. Review.
-
Computational Exploration of Bio-Degradation Patterns of Various Plastic Types.Polymers (Basel). 2023 Mar 20;15(6):1540. doi: 10.3390/polym15061540. Polymers (Basel). 2023. PMID: 36987320 Free PMC article.
-
Perspectives on the microorganisms with the potentials of PET-degradation.Front Microbiol. 2025 Mar 12;16:1541913. doi: 10.3389/fmicb.2025.1541913. eCollection 2025. Front Microbiol. 2025. PMID: 40143857 Free PMC article. Review.
-
Polyethylene Biodegradation by an Artificial Bacterial Consortium: Rhodococcus as a Competitive Plastisphere Species.Microbes Environ. 2024;39(3):ME24031. doi: 10.1264/jsme2.ME24031. Microbes Environ. 2024. PMID: 39085141 Free PMC article.
References
-
- Tyumina E.A., Bazhutin G.A., Vikhareva E.V., Selyaninov A.A., Ivshina I.B. Diclofenac as a factor in the change of Rhodococcus metabolism. IOP Conf. Ser. Mater. Sci. Eng. 2019;487:012027. doi: 10.1088/1757-899X/487/1/012027. - DOI
LinkOut - more resources
Full Text Sources

