3dDNA: A Computational Method of Building DNA 3D Structures
- PMID: 36144680
- PMCID: PMC9503956
- DOI: 10.3390/molecules27185936
3dDNA: A Computational Method of Building DNA 3D Structures
Abstract
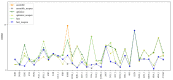
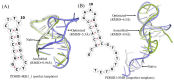
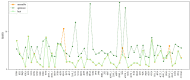
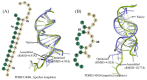
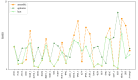
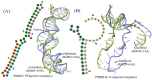
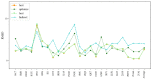
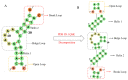
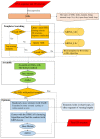
Considerable progress has been made in the prediction methods of 3D structures of RNAs. In contrast, no such methods are available for DNAs. The determination of 3D structures of the latter is also increasingly needed for understanding their functions and designing new DNA molecules. Since the number of experimental structures of DNA is limited at present, here, we propose a computational and template-based method, 3dDNA, which combines DNA and RNA template libraries to predict DNA 3D structures. It was benchmarked on three test sets with different numbers of chains, and the results show that 3dDNA can predict DNA 3D structures with a mean RMSD of about 2.36 Å for those with one or two chains and fewer than 4 Å with three or more chains.
Keywords: 3D structure prediction; 3D template libraries; 3dDNA; DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
3dRNA/DNA: 3D Structure Prediction from RNA to DNA.J Mol Biol. 2024 Sep 1;436(17):168742. doi: 10.1016/j.jmb.2024.168742. Epub 2024 Aug 7. J Mol Biol. 2024. PMID: 39237199
-
RNA 3D Structure Modeling by Combination of Template-Based Method ModeRNA, Template-Free Folding with SimRNA, and Refinement with QRNAS.Methods Mol Biol. 2016;1490:217-35. doi: 10.1007/978-1-4939-6433-8_14. Methods Mol Biol. 2016. PMID: 27665602
-
Advances in RNA 3D Structure Prediction.J Chem Inf Model. 2022 Dec 12;62(23):5862-5874. doi: 10.1021/acs.jcim.2c00939. Epub 2022 Nov 30. J Chem Inf Model. 2022. PMID: 36451090 Review.
-
A Method to Predict the 3D Structure of an RNA Scaffold.Methods Mol Biol. 2015;1316:1-11. doi: 10.1007/978-1-4939-2730-2_1. Methods Mol Biol. 2015. PMID: 25967048 Free PMC article.
-
RNA 3D Structure Prediction: Progress and Perspective.Molecules. 2023 Jul 20;28(14):5532. doi: 10.3390/molecules28145532. Molecules. 2023. PMID: 37513407 Free PMC article. Review.
Cited by
-
Computational Modeling of DNA 3D Structures: From Dynamics and Mechanics to Folding.Molecules. 2023 Jun 17;28(12):4833. doi: 10.3390/molecules28124833. Molecules. 2023. PMID: 37375388 Free PMC article. Review.
-
An Ensemble Docking Approach for Analyzing and Designing Aptamer Heterodimers Targeting VEGF165.Int J Mol Sci. 2024 Apr 5;25(7):4066. doi: 10.3390/ijms25074066. Int J Mol Sci. 2024. PMID: 38612876 Free PMC article.
-
Molecular Dynamics Simulation of Lipid Nanoparticles Encapsulating mRNA.Molecules. 2024 Sep 17;29(18):4409. doi: 10.3390/molecules29184409. Molecules. 2024. PMID: 39339404 Free PMC article.
-
Chromosome-scale genome assembly of Phyllanthus emblica L. 'Yingyu'.DNA Res. 2025 Mar 1;32(2):dsaf006. doi: 10.1093/dnares/dsaf006. DNA Res. 2025. PMID: 40070358 Free PMC article.
-
3dDNAscoreA: A scoring function for evaluation of DNA 3D structures.Biophys J. 2024 Sep 3;123(17):2696-2704. doi: 10.1016/j.bpj.2024.02.018. Epub 2024 Feb 26. Biophys J. 2024. PMID: 38409781
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

