In Silico Study towards Repositioning of FDA-Approved Drug Candidates for Anticoronaviral Therapy: Molecular Docking, Molecular Dynamics and Binding Free Energy Calculations
- PMID: 36144718
- PMCID: PMC9505381
- DOI: 10.3390/molecules27185988
In Silico Study towards Repositioning of FDA-Approved Drug Candidates for Anticoronaviral Therapy: Molecular Docking, Molecular Dynamics and Binding Free Energy Calculations
Abstract
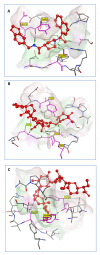
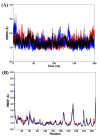
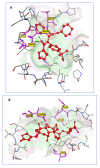
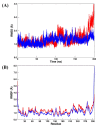
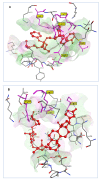
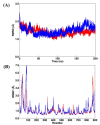
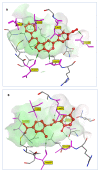
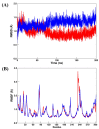
The SARS-CoV-2 targets were evaluated for a set of FDA-approved drugs using a combination of drug repositioning and rigorous computational modeling methodologies such as molecular docking and molecular dynamics (MD) simulations followed by binding free energy calculations. Six FDA-approved drugs including, Ouabain, Digitoxin, Digoxin, Proscillaridin, Salinomycin and Niclosamide with promising anti-SARS-CoV-2 activity were screened in silico against four SARS-CoV-2 proteins-papain-like protease (PLpro), RNA-dependent RNA polymerase (RdRp), SARS-CoV-2 main protease (Mpro), and adaptor-associated kinase 1 (AAK1)-in an attempt to define their promising targets. The applied computational techniques suggest that all the tested drugs exhibited excellent binding patterns with higher scores and stable complexes compared to the native protein cocrystallized inhibitors. Ouabain was suggested to act as a dual inhibitor for both PLpro and Mpro enzymes, while Digitoxin bonded perfectly to RdRp. In addition, Salinomycin targeted PLpro. Particularly, Niclosamide was found to target AAK1 with greater affinity compared to the reference drug. Our study provides comprehensive molecular-level insights for identifying or designing novel anti-COVID-19 drugs.
Keywords: anti-COVID-19; binding free energy; drug repositioning; molecular docking; molecular dynamic simulations.
Conflict of interest statement
There are no conflict to declare.
Figures

Similar articles
-
Identification of FDA approved drugs against SARS-CoV-2 RNA dependent RNA polymerase (RdRp) and 3-chymotrypsin-like protease (3CLpro), drug repurposing approach.Biomed Pharmacother. 2021 Jun;138:111544. doi: 10.1016/j.biopha.2021.111544. Epub 2021 Mar 31. Biomed Pharmacother. 2021. PMID: 34311539 Free PMC article.
-
Black tea bioactives as inhibitors of multiple targets of SARS-CoV-2 (3CLpro, PLpro and RdRp): a virtual screening and molecular dynamic simulation study.J Biomol Struct Dyn. 2022 Sep;40(15):7143-7166. doi: 10.1080/07391102.2021.1897679. Epub 2021 Mar 10. J Biomol Struct Dyn. 2022. PMID: 33715595
-
In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing.J Infect Public Health. 2020 Sep;13(9):1210-1223. doi: 10.1016/j.jiph.2020.06.016. Epub 2020 Jun 16. J Infect Public Health. 2020. PMID: 32561274 Free PMC article.
-
Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs.Precis Clin Med. 2021 Jan 18;4(1):1-16. doi: 10.1093/pcmedi/pbab001. eCollection 2021 Mar. Precis Clin Med. 2021. PMID: 33842834 Free PMC article. Review.
-
An Overview of SARS-CoV-2 Potential Targets, Inhibitors, and Computational Insights to Enrich the Promising Treatment Strategies.Curr Microbiol. 2024 May 11;81(7):169. doi: 10.1007/s00284-024-03671-3. Curr Microbiol. 2024. PMID: 38733424 Review.
Cited by
-
Computational Analysis of SAM Analogs as Methyltransferase Inhibitors of nsp16/nsp10 Complex from SARS-CoV-2.Int J Mol Sci. 2022 Nov 12;23(22):13972. doi: 10.3390/ijms232213972. Int J Mol Sci. 2022. PMID: 36430451 Free PMC article.
-
Computational Drug Design Strategies for Fighting the COVID-19 Pandemic.Adv Exp Med Biol. 2024;1457:199-214. doi: 10.1007/978-3-031-61939-7_11. Adv Exp Med Biol. 2024. PMID: 39283428
-
Advances and Challenges in Antiviral Development for Respiratory Viruses.Pathogens. 2024 Dec 31;14(1):20. doi: 10.3390/pathogens14010020. Pathogens. 2024. PMID: 39860981 Free PMC article. Review.
-
Main and papain-like proteases as prospective targets for pharmacological treatment of coronavirus SARS-CoV-2.RSC Adv. 2023 Dec 6;13(50):35500-35524. doi: 10.1039/d3ra06479d. eCollection 2023 Nov 30. RSC Adv. 2023. PMID: 38077980 Free PMC article. Review.
-
Potential Anti-SARS-CoV-2 Molecular Strategies.Molecules. 2023 Feb 24;28(5):2118. doi: 10.3390/molecules28052118. Molecules. 2023. PMID: 36903364 Free PMC article.
References
-
- WHO Coronavirus Disease (COVID-19) [(accessed on 4 January 2022)]. Available online: https://covid19.who.int/
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

