A Label-Free Fluorescence Aptasensor Based on G-Quadruplex/Thioflavin T Complex for the Detection of Trypsin
- PMID: 36144829
- PMCID: PMC9503660
- DOI: 10.3390/molecules27186093
A Label-Free Fluorescence Aptasensor Based on G-Quadruplex/Thioflavin T Complex for the Detection of Trypsin
Abstract
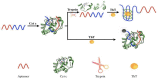
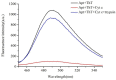
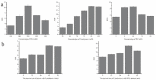
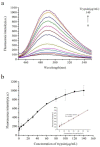
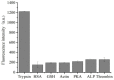
A novel, label-free fluorescent assay has been developed for the detection of trypsin by using thioflavin T as a fluorescent probe. A specific DNA aptamer can be combined by adding cytochrome c. Trypsin hydrolyzes the cytochrome c into small peptide fragments, exposing the G-quadruplex part of DNA aptamer, which has a high affinity for thioflavin T, which then enhances the fluorescence intensity. In the absence of trypsin, the fluorescence intensity was inhibited as the combination of cytochrome c and the DNA aptamer impeded thioflavin T's binding. Thus, the fluorescent biosensor showed a linear relationship from 0.2 to 60 μg/mL with a detection limit of 0.2 μg/mL. Furthermore, the proposed method was also successfully employed for determining trypsin in biological samples. This method is simple, rapid, cheap, and selective and possesses great potential for the detection of trypsin in bioanalytical and biological samples and medical diagnoses.
Keywords: DNA aptamer; cytochrome c; label-free; thioflavin T; trypsin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
G-quadruplex specific thioflavin T-based label-free fluorescence aptasensor for rapid detection of tetracycline.Spectrochim Acta A Mol Biomol Spectrosc. 2020 Sep 5;238:118406. doi: 10.1016/j.saa.2020.118406. Epub 2020 Apr 23. Spectrochim Acta A Mol Biomol Spectrosc. 2020. PMID: 32387918
-
A facile label-free G-quadruplex based fluorescent aptasensor method for rapid detection of ATP.Spectrochim Acta A Mol Biomol Spectrosc. 2017 Mar 15;175:164-167. doi: 10.1016/j.saa.2016.12.033. Epub 2016 Dec 21. Spectrochim Acta A Mol Biomol Spectrosc. 2017. PMID: 28038373
-
Novel strategy to improve the sensing performances of split ATP aptamer based fluorescent indicator displacement assay through enhanced molecular recognition.Biosens Bioelectron. 2019 Jun 1;134:36-41. doi: 10.1016/j.bios.2019.03.047. Epub 2019 Mar 27. Biosens Bioelectron. 2019. PMID: 30954924
-
Label-Free G-Quadruplex Aptamer Fluorescence Assay for Ochratoxin A Using a Thioflavin T Probe.Toxins (Basel). 2018 May 12;10(5):198. doi: 10.3390/toxins10050198. Toxins (Basel). 2018. PMID: 29757205 Free PMC article.
-
A rapid fluorometric method for determination of aflatoxin B1 in plant-derived food by using a thioflavin T-based aptasensor.Mikrochim Acta. 2019 Mar 4;186(4):214. doi: 10.1007/s00604-019-3325-9. Mikrochim Acta. 2019. PMID: 30830273
Cited by
-
Telomere and subtelomere high polymorphism might contribute to the specificity of homologous recognition and pairing during meiosis in barley in the context of breeding.BMC Genomics. 2023 Oct 26;24(1):642. doi: 10.1186/s12864-023-09738-y. BMC Genomics. 2023. PMID: 37884878 Free PMC article.
References
-
- Lasher D., Szabó A., Masamune A., Chen J.M., Xiao X., Whitcomb D.C., Barmada M.M., Ewers M., Ruffert C., Paliwal S., et al. Protease-Sensitive Pancreatic Lipase Variants Are Associated With Early Onset Chronic Pancreatitis. Am. J. Gastroenterol. 2019;114:974–983. doi: 10.14309/ajg.0000000000000051. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

