Strain Variation Based on Spike Glycoprotein Gene of SARS-CoV-2 in Kuwait from 2020 to 2021
- PMID: 36145416
- PMCID: PMC9505955
- DOI: 10.3390/pathogens11090985
Strain Variation Based on Spike Glycoprotein Gene of SARS-CoV-2 in Kuwait from 2020 to 2021
Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19), which was first identified in Wuhan, China, in December 2019. With the global transmission of the virus, many SARS-CoV-2 variants have emerged due to the alterations of the spike glycoprotein. Therefore, the S glycoprotein encoding gene has widely been used for the molecular analysis of SARS-Co-2 due to its features affecting antigenicity and immunogenicity. We analyzed the S gene sequences of 35 SARS-CoV-2 isolates in Kuwait from March 2020 to February 2021 using the Sanger method and MinION nanopore technology to confirm novel nucleotide alterations. Our results show that the Kuwaiti strains from clade 19A and B were the dominant variants early in the pandemic, while clade 20I (Alpha, V1) was the dominant variant from February 2021 onward. Besides the known mutations, 21 nucleotide deletions in the S glycoprotein in one Kuwaiti strain were detected, which might reveal a recombinant SARS-CoV-2 with the defective viral genome (DVG). This study emphasizes the importance of closely perceiving the emerging clades with these mutations during this continuous pandemic as some may influence the specificity of diagnostic tests, such as RT-PCR and even vaccine design directing these positions.
Keywords: Kuwait; SARS-CoV-2; spike glycoprotein; strain variation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
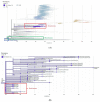


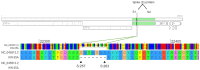
References
-
- Ralph R., Lew J., Zeng T., Francis M., Xue B., Roux M., Ostadgavahi A.T., Rubino S., Dawe N.J., Al-Ahdal M.N., et al. 2019-NCoV (Wuhan Virus), a Novel Coronavirus: Human-to-Human Transmission, Travel-Related Cases, and Vaccine Readiness. J. Infect. Dev. Ctries. 2020;14:3–17. doi: 10.3855/jidc.12425. - DOI - PubMed
-
- COVID-19 Map—Johns Hopkins Coronavirus Resource Center. [(accessed on 26 April 2021)]. Available online: https://coronavirus.jhu.edu/map.html.
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. - PMC - PubMed
-
- Mohammad Lokman S., Rasheduzzaman M., Salauddin A., Barua R., Yeasmin Tanzina A., Hasan Rumi M., Imran Hossain M., Zonaed Siddiki A., Mannan A., Mahbub Hasan M. Exploring the Genomic and Proteomic Variations of SARS-CoV-2 Spike Glycoprotein: A Computational Biology Approach. Infect. Genet. Evol. 2020;84:104389. doi: 10.1016/j.meegid.2020.104389. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

