Characterization of a Near Full-Length Hepatitis E Virus Genome of Subtype 3c Generated from Naturally Infected South African Backyard Pigs
- PMID: 36145462
- PMCID: PMC9506134
- DOI: 10.3390/pathogens11091030
Characterization of a Near Full-Length Hepatitis E Virus Genome of Subtype 3c Generated from Naturally Infected South African Backyard Pigs
Abstract
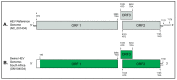
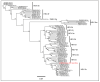
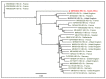
Eight genotypes of the hepatitis E virus (Orthohepevirus A; HEV) designated HEV-1 to HEV-8 have been reported from various mammalian hosts. Notably, domestic pigs and wild boars are the natural reservoirs of HEV-3 and HEV-4 genotypes with zoonotic propensity. Since HEV infection in domestic pigs is usually subclinical, it may remain undetected, facilitating zoonotic spillover of HEV to the exposed human populations. A previous study from our group in 2021, using deep sequencing of a pooled saliva sample, generated various swine enteric virus genomes, including a near full-length swine HEV genome (7040 nt; 97.7% genome coverage) from five-month-old grower pigs at a backyard pig farm in the uMgungundlovu District, KwaZulu-Natal, South Africa. In the present study, we describe the further characterization, including genotyping and subtyping of the swine HEV isolate using phylogenetics and ‘HEVnet Typing Tool’. Our analyses confirmed that the South African swine HEV genome characterized in this study belonged to HEV genotype 3 subtype 3c (HEV-3c). While HEV-3c infections in domestic pigs have been previously reported from Brazil, Germany, Italy, and the Netherlands, they only generated partial genome sequences of open reading frame 1 (ORF1) and/or ORF2. To our knowledge, this is the first near full-length swine HEV-3c genome generated from naturally infected domestic pigs (Sus scrofa domesticus) in South Africa. However, due to the gap in the information on the HEV-3c genome sequences in various geographical locations worldwide, including South Africa, the epidemiology of the South African swine HEV genome characterized in this study remains inconclusive. Molecular and genomic surveillance of HEV in domestic pig populations in South Africa would be useful to determine their prevalence, circulating subtypes, and zoonosis risk.
Keywords: HEV zoonosis; HEV-3c; Illumina sequencing; Sus scrofa domesticus; backyard pig farm; hepatitis E virus; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Metagenomic Analysis of RNA Fraction Reveals the Diversity of Swine Oral Virome on South African Backyard Swine Farms in the uMgungundlovu District of KwaZulu-Natal Province.Pathogens. 2022 Aug 17;11(8):927. doi: 10.3390/pathogens11080927. Pathogens. 2022. PMID: 36015047 Free PMC article.
-
Prevalence and phylogenetic analysis of hepatitis E virus in pigs, wild boars, roe deer, red deer and moose in Lithuania.Acta Vet Scand. 2018 Feb 23;60(1):13. doi: 10.1186/s13028-018-0367-7. Acta Vet Scand. 2018. PMID: 29471843 Free PMC article.
-
Molecular detection and phylogenetic analysis of hepatitis E virus strains circulating in wild boars in south-central Italy.Transbound Emerg Dis. 2018 Feb;65(1):e25-e31. doi: 10.1111/tbed.12661. Epub 2017 May 11. Transbound Emerg Dis. 2018. PMID: 28497542
-
Hepatitis E Virus of Subtype 3a in a Pig Farm, South-Eastern France.Zoonoses Public Health. 2015 Dec;62(8):593-8. doi: 10.1111/zph.12211. Epub 2015 Jun 23. Zoonoses Public Health. 2015. PMID: 26102074
-
A Systematic Review and Meta-Analysis on Hepatitis E Virus Detection in Farmed Ruminants.Pathogens. 2023 Apr 2;12(4):550. doi: 10.3390/pathogens12040550. Pathogens. 2023. PMID: 37111437 Free PMC article. Review.
Cited by
-
Overview of Diagnostic Methods, Disease Prevalence and Transmission of Mpox (Formerly Monkeypox) in Humans and Animal Reservoirs.Microorganisms. 2023 Apr 30;11(5):1186. doi: 10.3390/microorganisms11051186. Microorganisms. 2023. PMID: 37317160 Free PMC article. Review.
-
Water-Based Epidemiological Investigation of Hepatitis E Virus in South Africa.Food Environ Virol. 2024 Sep;16(3):338-350. doi: 10.1007/s12560-024-09596-1. Epub 2024 Apr 13. Food Environ Virol. 2024. PMID: 38613652 Free PMC article.
-
Hepatitis E Seroprevalence and Detection of Genotype 3 Strains in Domestic Pigs from Sierra Leone Collected in 2016 and 2017.Viruses. 2024 Apr 3;16(4):558. doi: 10.3390/v16040558. Viruses. 2024. PMID: 38675900 Free PMC article.
-
Zoonotic and Food-Related Hazards Due to Hepatitis A and E in Africa: A Systematic Review and Meta-Analysis.Environ Health Insights. 2024 Nov 21;18:11786302241299370. doi: 10.1177/11786302241299370. eCollection 2024. Environ Health Insights. 2024. PMID: 39575136 Free PMC article. Review.
References
-
- Henritzi D., Petric P.P., Lewis N.S., Graaf A., Pessia A., Starick E., Breithaupt A., Strebelow G., Luttermann C., Parker L.M.K., et al. Surveillance of European Domestic Pig Populations Identifies an Emerging Reservoir of Potentially Zoonotic Swine Influenza A Viruses. Cell Host Microbe. 2020;28:614–627.e616. doi: 10.1016/j.chom.2020.07.006. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

