Structure of Lacticaseicin 30 and Its Engineered Variants Revealed an Interplay between the N-Terminal and C-Terminal Regions in the Activity against Gram-Negative Bacteria
- PMID: 36145669
- PMCID: PMC9505257
- DOI: 10.3390/pharmaceutics14091921
Structure of Lacticaseicin 30 and Its Engineered Variants Revealed an Interplay between the N-Terminal and C-Terminal Regions in the Activity against Gram-Negative Bacteria
Abstract
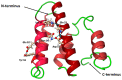
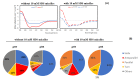
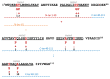
Lacticaseicin 30 is one of the five bacteriocins produced by the Gram-positive Lacticaseibacillus paracasei CNCM I-5369. This 111 amino acid bacteriocin is noteworthy for being active against Gram-negative bacilli including Escherichia coli strains resistant to colistin. Prediction of the lacticaseicin 30 structure using the Alphafold2 pipeline revealed a largely helical structure including five helix segments, which was confirmed by circular dichroism. To identify the structural requirements of the lacticaseicin 30 activity directed against Gram-negative bacilli, a series of variants, either shortened or containing point mutations, was heterologously produced in Escherichia coli and assayed for their antibacterial activity against a panel of target strains including Gram-negative bacteria and the Gram-positive Listeria innocua. Lacticaseicin 30 variants comprising either the N-terminal region (amino acids 1 to 39) or the central and C-terminal regions (amino acids 40 to 111) were prepared. Furthermore, mutations were introduced by site-directed mutagenesis to obtain ten bacteriocin variants E6G, T7P, E32G, T33P, T52P, D57G, A74P, Y78S, Y93S and A97P. Compared to lacticaseicin 30, the anti-Gram-negative activity of the N-terminal peptide and variants E32G, T33P and D57G remained almost unchanged, while that of the C-terminal peptide and variants E6G, T7P, T52P, A74P, Y78S, Y93S and A97P was significantly altered. Finally, the N-terminal region was further shortened to keep only the first 20 amino acid part that was predicted to include the first helix. The anti-Gram-negative activity of this truncated peptide was completely abolished. Overall, this study shows that activity of lacticaseicin 30, one of the rare Gram-positive bacteriocins inhibiting Gram-negative bacteria, requires at least two helices in the N-terminal region and that the C-terminal region carries amino acids playing a role in modulation of the activity. Taken together, these data will help to design forthcoming variants of lacticaseicin 30 as promising therapeutic agents to treat infections caused by Gram-negative bacilli.
Keywords: Escherichia coli; antimicrobial activity; helical conformation; structure-activity relationship.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Lacticaseicin 30 and Colistin as a Promising Antibiotic Formulation against Gram-Negative β-Lactamase-Producing Strains and Colistin-Resistant Strains.Antibiotics (Basel). 2021 Dec 24;11(1):20. doi: 10.3390/antibiotics11010020. Antibiotics (Basel). 2021. PMID: 35052897 Free PMC article.
-
Bacteriocins of gram-positive bacteria.Microbiol Rev. 1995 Jun;59(2):171-200. doi: 10.1128/mr.59.2.171-200.1995. Microbiol Rev. 1995. PMID: 7603408 Free PMC article. Review.
-
Peptide-bacteria interactions using engineered surface-immobilized peptides from class IIa bacteriocins.Langmuir. 2013 Mar 26;29(12):4048-56. doi: 10.1021/la3041743. Epub 2013 Mar 14. Langmuir. 2013. PMID: 23445325
-
Covalent structure, synthesis, and structure-function studies of mesentericin Y 105(37), a defensive peptide from gram-positive bacteria Leuconostoc mesenteroides.J Biol Chem. 1996 Jun 14;271(24):14421-9. doi: 10.1074/jbc.271.24.14421. J Biol Chem. 1996. PMID: 8662868
-
Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by gram-positive bacteria.Curr Pharm Biotechnol. 2009 Jan;10(1):19-37. doi: 10.2174/138920109787048661. Curr Pharm Biotechnol. 2009. PMID: 19149588 Review.
Cited by
-
AlphaFold2 in biomedical research: facilitating the development of diagnostic strategies for disease.Front Mol Biosci. 2024 Jul 30;11:1414916. doi: 10.3389/fmolb.2024.1414916. eCollection 2024. Front Mol Biosci. 2024. PMID: 39139810 Free PMC article. Review.
-
The Editorial Position on 'Recent Advances in Multifunctional Antimicrobial Peptides as Preclinical Therapeutic Studies and Clinical Future Applications'.Pharmaceutics. 2023 Sep 26;15(10):2383. doi: 10.3390/pharmaceutics15102383. Pharmaceutics. 2023. PMID: 37896143 Free PMC article.
-
Enterocin DD14 can inhibit the infection of eukaryotic cells with enveloped viruses.Arch Microbiol. 2024 May 20;206(6):269. doi: 10.1007/s00203-024-04002-7. Arch Microbiol. 2024. PMID: 38767708
References
-
- O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance; London, UK: 2016.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

