Effects of Prior Infection with SARS-CoV-2 on B Cell Receptor Repertoire Response during Vaccination
- PMID: 36146555
- PMCID: PMC9506540
- DOI: 10.3390/vaccines10091477
Effects of Prior Infection with SARS-CoV-2 on B Cell Receptor Repertoire Response during Vaccination
Abstract
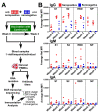
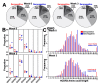
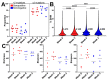
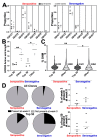
Understanding the B cell response to SARS-CoV-2 vaccines is a high priority. High-throughput sequencing of the B cell receptor (BCR) repertoire allows for dynamic characterization of B cell response. Here, we sequenced the BCR repertoire of individuals vaccinated by the Pfizer SARS-CoV-2 mRNA vaccine. We compared BCR repertoires of individuals with previous COVID-19 infection (seropositive) to individuals without previous infection (seronegative). We discovered that vaccine-induced expanded IgG clonotypes had shorter heavy-chain complementarity determining region 3 (HCDR3), and for seropositive individuals, these expanded clonotypes had higher somatic hypermutation (SHM) than seronegative individuals. We uncovered shared clonotypes present in multiple individuals, including 28 clonotypes present across all individuals. These 28 shared clonotypes had higher SHM and shorter HCDR3 lengths compared to the rest of the BCR repertoire. Shared clonotypes were present across both serotypes, indicating convergent evolution due to SARS-CoV-2 vaccination independent of prior viral exposure.
Keywords: B cell receptor heavy-chain sequencing; SARS-CoV-2; vaccine response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Comparative global B cell receptor repertoire difference induced by SARS-CoV-2 infection or vaccination via single-cell V(D)J sequencing.Emerg Microbes Infect. 2022 Dec;11(1):2007-2020. doi: 10.1080/22221751.2022.2105261. Emerg Microbes Infect. 2022. PMID: 35899581 Free PMC article.
-
Imprints of somatic hypermutation on B-cell receptor immunoglobulins post-infection versus post-vaccination against SARS-CoV-2.Immunohorizons. 2025 May 30;9(7):vlaf021. doi: 10.1093/immhor/vlaf021. Immunohorizons. 2025. PMID: 40489958 Free PMC article.
-
Analysis of B Cell Receptor Repertoires Reveals Key Signatures of the Systemic B Cell Response after SARS-CoV-2 Infection.J Virol. 2022 Feb 23;96(4):e0160021. doi: 10.1128/JVI.01600-21. Epub 2021 Dec 8. J Virol. 2022. PMID: 34878902 Free PMC article.
-
Utility of Bulk T-Cell Receptor Repertoire Sequencing Analysis in Understanding Immune Responses to COVID-19.Diagnostics (Basel). 2022 May 13;12(5):1222. doi: 10.3390/diagnostics12051222. Diagnostics (Basel). 2022. PMID: 35626377 Free PMC article. Review.
-
The shape of the lymphocyte receptor repertoire: lessons from the B cell receptor.Front Immunol. 2013 Sep 2;4:263. doi: 10.3389/fimmu.2013.00263. Front Immunol. 2013. PMID: 24032032 Free PMC article. Review.
Cited by
-
A humanized mouse that mounts mature class-switched, hypermutated and neutralizing antibody responses.Nat Immunol. 2024 Aug;25(8):1489-1506. doi: 10.1038/s41590-024-01880-3. Epub 2024 Jun 25. Nat Immunol. 2024. PMID: 38918608 Free PMC article.
-
Naive and Memory B Cell BCR Repertoires in Individuals Immunized with an Inactivated SARS-CoV-2 Vaccine.Vaccines (Basel). 2025 Apr 8;13(4):393. doi: 10.3390/vaccines13040393. Vaccines (Basel). 2025. PMID: 40333337 Free PMC article.
-
A comprehensive immune repertoire signature distinguishes pulmonary infiltration in SARS-CoV-2 Omicron variant infection.Front Immunol. 2024 Dec 17;15:1486352. doi: 10.3389/fimmu.2024.1486352. eCollection 2024. Front Immunol. 2024. PMID: 39742285 Free PMC article.
-
Revaccination in Age-Risk Groups with Sputnik V Is Immunologically Effective and Depends on the Initial Neutralizing SARS-CoV-2 IgG Antibodies Level.Vaccines (Basel). 2022 Dec 30;11(1):90. doi: 10.3390/vaccines11010090. Vaccines (Basel). 2022. PMID: 36679936 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

