Human Fc-Conjugated Receptor Binding Domain-Based Recombinant Subunit Vaccines with Short Linker Induce Potent Neutralizing Antibodies against Multiple SARS-CoV-2 Variants
- PMID: 36146579
- PMCID: PMC9505662
- DOI: 10.3390/vaccines10091502
Human Fc-Conjugated Receptor Binding Domain-Based Recombinant Subunit Vaccines with Short Linker Induce Potent Neutralizing Antibodies against Multiple SARS-CoV-2 Variants
Abstract
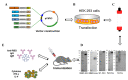
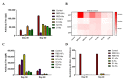
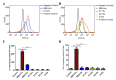
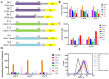
The coronavirus disease-19 (COVID-19) pandemic has been ongoing since December 2019, with more than 6.3 million deaths reported globally as of August 2022. Despite the success of several SARS-CoV-2 vaccines, the rise in variants, some of which are resistant to the effects of vaccination, highlights the need for a so-called pan-coronavirus (universal) vaccine. Here, we performed an immunogenicity comparison of prototype vaccines containing spike protein receptor-binding domain (RBD) residues 319-541, or spike protein regions S1, S2 and S fused to a histidine-tagged or human IgG1 Fc (hFC) fragment with either a longer (six residues) or shorter (three residues) linker. While all recombinant protein vaccines developed were effective in eliciting humoral immunity, the RBD-hFc vaccine was able to generate a potent neutralizing antibody response as well as a cellular immune response. We then compared the effects of recombinant protein length and linker size on immunogenicity in vivo. We found that a longer recombinant RBD protein (residues 319-583; RBD-Plus-hFc) containing a small alanine linker (AAA) was able to trigger long-lasting, high-titer neutralizing antibodies in mice. Finally, we evaluated cross-neutralization of wild-type and mutant RBD-Plus-hFc vaccines against wild-type, Alpha, Beta, Delta and Omicron SARS-CoV-2 variants. Significantly, at the same antigen dose, wild-type RBD-Plus-hFc immune sera induced broadly neutralizing antibodies against wild-type, Alpha, Beta, Delta and Omicron variants. Taken together, our findings provide valuable information for the continued development of recombinant protein-based SARS-CoV-2 vaccines and a basic foundation for booster vaccinations to avoid reinfection with SARS-CoV-2 variants.
Keywords: RBD; SARS-CoV-2 variants; recombinant protein subunit vaccines; spike protein.
Conflict of interest statement
The recombinant protein vaccines used in this paper have been patented by Guangdong Keguanda Biomedical Technology Co., Ltd. The other authors declare that they have no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

