Comprehensive Characterization of Immune Landscape Based on Tumor Microenvironment for Oral Squamous Cell Carcinoma Prognosis
- PMID: 36146599
- PMCID: PMC9505673
- DOI: 10.3390/vaccines10091521
Comprehensive Characterization of Immune Landscape Based on Tumor Microenvironment for Oral Squamous Cell Carcinoma Prognosis
Abstract
Objective: This study aims to identify an immune-related signature to predict clinical outcomes of oral squamous cell carcinoma (OSCC) patients.
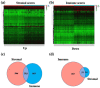
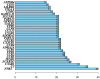
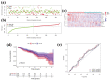
Methods: Gene transcriptome data of both tumor and normal tissues from OSCC and the corresponding clinical information were downloaded from The Cancer Genome Atlas (TCGA). Tumor Immune Estimation Resource algorithm (ESTIMATE) was used to calculate the immune/stromal-related scores. The immune/stromal scores and associated clinical characteristics of OSCC patients were evaluated. Univariate Cox proportional hazards regression analyses, least absolute shrinkage, and selection operator (LASSO) and receiver operating characteristic (ROC) curve analyses were performed to assess the prognostic prediction capacity. Gene Set Enrichment Analysis (GSEA) and Gene Ontology (GO) function annotation were used to analysis the functions of TME-related genes.
Results: Eleven predictor genes were identified in the immune-related signature and overall survival (OS) in the high-risk group was significantly shorter than in the low-risk group. An ROC analysis showed the TME-related signature could predict the total OS of OSCC patients. Moreover, GSEA and GO function annotation proved that immunity and immune-related pathways were mainly enriched in the high-risk group.
Conclusions: We identified an immune-related signature that was closely correlated with the prognosis and immune response of OSCC patients. This signature may have important implications for improving the clinical survival rate of OSCC patients and provide a potential strategy for cancer immunotherapy.
Keywords: immunotherapy; oral squamous cell carcinoma; prognosis; signature; survival.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Yang Z., Yan G., Zheng L., Gu W., Liu F., Chen W., Cui X., Wang Y., Yang Y., Chen X., et al. YKT6, as a potential predictor of prognosis and immunotherapy response for oral squamous cell carcinoma, is related to cell invasion, metastasis, and CD8+ T cell infiltration. Oncoimmunology. 2021;10:1938890. doi: 10.1080/2162402X.2021.1938890. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

