The Genetic Characterization of the First Detected Bat Coronaviruses in Poland Revealed SARS-Related Types and Alphacoronaviruses
- PMID: 36146721
- PMCID: PMC9501061
- DOI: 10.3390/v14091914
The Genetic Characterization of the First Detected Bat Coronaviruses in Poland Revealed SARS-Related Types and Alphacoronaviruses
Abstract
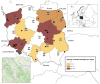
Bats are a major global reservoir of alphacoronaviruses (alphaCoVs) and betaCoVs. Attempts to discover the causative agents of COVID-19 and SARS have revealed horseshoe bats (Rhinolophidae) to be the most probable source of the virus. We report the first detection of bat coronaviruses (BtCoVs) in insectivorous bats in Poland and highlight SARS-related coronaviruses found in Rhinolophidae bats. The study included 503 (397 oral swabs and 106 fecal) samples collected from 20 bat species. Genetically diverse BtCoVs (n = 20) of the Alpha- and Betacoronavirus genera were found in fecal samples of two bat species. SARS-related CoVs were in 18 out of 58 lesser horseshoe bat (Rhinolophus hipposideros) samples (31%, 95% CI 20.6-43.8), and alphaCoVs were in 2 out of 55 Daubenton's bat (Myotis daubentonii) samples (3.6%, 95% CI 0.6-12.3). The overall BtCoV prevalence was 4.0% (95% CI 2.6-6.1). High identity was determined for BtCoVs isolated from European M. daubentonii and R. hipposideros bats. The detection of SARS-related and alphaCoVs in Polish bats with high phylogenetic relatedness to reference BtCoVs isolated in different European countries but from the same species confirms their high host restriction. Our data elucidate the molecular epidemiology, prevalence, and geographic distribution of coronaviruses and particularly SARS-related types in the bat population.
Keywords: Poland; SARS-related CoV; bats; coronaviruses; phylogenetics; prevalence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Novel Alphacoronaviruses and Paramyxoviruses Cocirculate with Type 1 and Severe Acute Respiratory System (SARS)-Related Betacoronaviruses in Synanthropic Bats of Luxembourg.Appl Environ Microbiol. 2017 Aug 31;83(18):e01326-17. doi: 10.1128/AEM.01326-17. Print 2017 Sep 15. Appl Environ Microbiol. 2017. PMID: 28710271 Free PMC article.
-
SARS-CoV related Betacoronavirus and diverse Alphacoronavirus members found in western old-world.Virology. 2018 Apr;517:88-97. doi: 10.1016/j.virol.2018.01.014. Epub 2018 Feb 23. Virology. 2018. PMID: 29482919 Free PMC article.
-
The Role of Molossidae and Vespertilionidae in Shaping the Diversity of Alphacoronaviruses in the Americas.Microbiol Spectr. 2022 Dec 21;10(6):e0314322. doi: 10.1128/spectrum.03143-22. Epub 2022 Oct 12. Microbiol Spectr. 2022. PMID: 36222689 Free PMC article.
-
Global Epidemiology of Bat Coronaviruses.Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. Viruses. 2019. PMID: 30791586 Free PMC article. Review.
-
[Etiology of epidemic outbreaks COVID-19 on Wuhan, Hubei province, Chinese People Republic associated with 2019-nCoV (Nidovirales, Coronaviridae, Coronavirinae, Betacoronavirus, Subgenus Sarbecovirus): lessons of SARS-CoV outbreak.].Vopr Virusol. 2020;65(1):6-15. doi: 10.36233/0507-4088-2020-65-1-6-15. Vopr Virusol. 2020. PMID: 32496715 Review. Russian.
Cited by
-
A comprehensive dataset of animal-associated sarbecoviruses.Sci Data. 2023 Oct 7;10(1):681. doi: 10.1038/s41597-023-02558-5. Sci Data. 2023. PMID: 37805633 Free PMC article.
-
Circulation of SARS-CoV-Related Coronaviruses and Alphacoronaviruses in Bats from Croatia.Microorganisms. 2023 Apr 7;11(4):959. doi: 10.3390/microorganisms11040959. Microorganisms. 2023. PMID: 37110383 Free PMC article.
-
Prevalence of coronaviruses in European bison (Bison bonasus) in Poland.Sci Rep. 2024 Jun 5;14(1):12928. doi: 10.1038/s41598-024-63717-1. Sci Rep. 2024. PMID: 38839918 Free PMC article.
-
Lack of detection of SARS-CoV-2 in British wildlife 2020-21 and first description of a stoat (Mustela erminea) Minacovirus.J Gen Virol. 2023 Dec;104(12):001917. doi: 10.1099/jgv.0.001917. J Gen Virol. 2023. PMID: 38059490 Free PMC article.
References
-
- Ciechanowski M. Tom 3 Część 3 Zoologia: Ssaki. Polskie Wydawnictwo Naukowe (PWN); Warsaw, Poland: 2020. Rząd: Nietoperze–Chiroptera (Order: Chiroptera (bats)–in Polish) pp. 236–304.
-
- Taylor M. Bats: An Illustrated Guide to All Species. Smithsonian Books; Washington, DC, USA: 2019.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

