Phage Diving: An Exploration of the Carcharhinid Shark Epidermal Virome
- PMID: 36146775
- PMCID: PMC9500685
- DOI: 10.3390/v14091969
Phage Diving: An Exploration of the Carcharhinid Shark Epidermal Virome
Abstract
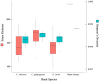
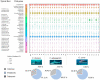
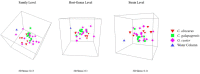
The epidermal microbiome is a critical element of marine organismal immunity, but the epidermal virome of marine organisms remains largely unexplored. The epidermis of sharks represents a unique viromic ecosystem. Sharks secrete a thin layer of mucus which harbors a diverse microbiome, while their hydrodynamic dermal denticles simultaneously repel environmental microbes. Here, we sampled the virome from the epidermis of three shark species in the family Carcharhinidae: the genetically and morphologically similar Carcharhinus obscurus (n = 6) and Carcharhinus galapagensis (n = 10) and the outgroup Galeocerdo cuvier (n = 15). Virome taxonomy was characterized using shotgun metagenomics and compared with a suite of multivariate analyses. All three sharks retain species-specific but highly similar epidermal viromes dominated by uncharacterized bacteriophages which vary slightly in proportional abundance within and among shark species. Intraspecific variation was lower among C. galapagensis than among C. obscurus and G. cuvier. Using both the annotated and unannotated reads, we were able to determine that the Carcharhinus galapagensis viromes were more similar to that of G. cuvier than they were to that of C. obscurus, suggesting that behavioral niche may be a more prominent driver of virome than host phylogeny.
Keywords: bacteriophage; carcharhinid; denticle; epidermis; metagenomics; shark; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Observations of sharks (Elasmobranchii) at Europa Island, a remote marine protected area important for shark conservation in the southern Mozambique Channel.PLoS One. 2021 Oct 5;16(10):e0253867. doi: 10.1371/journal.pone.0253867. eCollection 2021. PLoS One. 2021. PMID: 34610033 Free PMC article.
-
Vertical movement patterns and ontogenetic niche expansion in the tiger shark, Galeocerdo cuvier.PLoS One. 2015 Jan 28;10(1):e0116720. doi: 10.1371/journal.pone.0116720. eCollection 2015. PLoS One. 2015. PMID: 25629732 Free PMC article.
-
Feeding ecology and trophic comparisons of six shark species in a coastal ecosystem off southern Brazil.J Fish Biol. 2014 Aug;85(2):246-63. doi: 10.1111/jfb.12417. Epub 2014 Jun 12. J Fish Biol. 2014. PMID: 24919949
-
On the complexity of shark bite wounds: From associated bacteria to trauma management and wound repair.J Trauma Acute Care Surg. 2018 Aug;85(2):398-405. doi: 10.1097/TA.0000000000001920. J Trauma Acute Care Surg. 2018. PMID: 29613948 Review.
-
Bacteriophages of the Order Crassvirales: What Do We Currently Know about This Keystone Component of the Human Gut Virome?Biomolecules. 2023 Mar 24;13(4):584. doi: 10.3390/biom13040584. Biomolecules. 2023. PMID: 37189332 Free PMC article. Review.
Cited by
-
Phables: from fragmented assemblies to high-quality bacteriophage genomes.Bioinformatics. 2023 Oct 3;39(10):btad586. doi: 10.1093/bioinformatics/btad586. Bioinformatics. 2023. PMID: 37738590 Free PMC article.
-
Host interactions of novel Crassvirales species belonging to multiple families infecting bacterial host, Bacteroides cellulosilyticus WH2.bioRxiv [Preprint]. 2023 Jul 26:2023.03.05.531146. doi: 10.1101/2023.03.05.531146. bioRxiv. 2023. Update in: Microb Genom. 2023 Sep;9(9). doi: 10.1099/mgen.0.001100. PMID: 36945541 Free PMC article. Updated. Preprint.
-
Host interactions of novel Crassvirales species belonging to multiple families infecting bacterial host, Bacteroides cellulosilyticus WH2.Microb Genom. 2023 Sep;9(9):001100. doi: 10.1099/mgen.0.001100. Microb Genom. 2023. PMID: 37665209 Free PMC article.
-
Phables: from fragmented assemblies to high-quality bacteriophage genomes.bioRxiv [Preprint]. 2023 Sep 11:2023.04.04.535632. doi: 10.1101/2023.04.04.535632. bioRxiv. 2023. Update in: Bioinformatics. 2023 Oct 3;39(10):btad586. doi: 10.1093/bioinformatics/btad586. PMID: 37066369 Free PMC article. Updated. Preprint.
-
Identification and characterization of a polyomavirus in the thornback skate (Raja clavata).Virol J. 2023 Aug 24;20(1):190. doi: 10.1186/s12985-023-02149-1. Virol J. 2023. PMID: 37620878 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

