Epidemiological and Genomic Characterisation of Middelburg and Sindbis Alphaviruses Identified in Horses with Febrile and Neurological Infections, South Africa (2014-2018)
- PMID: 36146819
- PMCID: PMC9501102
- DOI: 10.3390/v14092013
Epidemiological and Genomic Characterisation of Middelburg and Sindbis Alphaviruses Identified in Horses with Febrile and Neurological Infections, South Africa (2014-2018)
Abstract
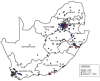
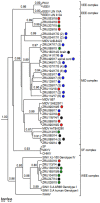
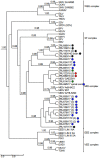
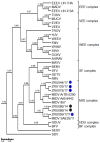
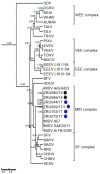
Although Old World alphaviruses, Middelburg- (MIDV) and Sindbis virus (SINV), have previously been detected in horses and wildlife with neurologic disease in South Africa, the pathogenesis and clinical presentation of MIDV and SINV infections in animals are not well documented. Clinical samples from horses across South Africa with acute or fatal neurologic and febrile infections submitted between 2014-2018 were investigated. In total, 69/1084 (6.36%) and 11/1084 (1.01%) horses tested positive for MIDV and SINV, respectively, by real-time reverse transcription (RT) PCR. Main signs/outcomes for MIDV (n = 69): 73.91% neurological, 75.36% fever, 28.99% icterus and anorexia, respectively, 8.70% fatalities; SINV (n = 11): 54.54% neurological, 72.73% fever, 36.36% anorexia and 18.18% fatalities. MIDV cases peaked in the late summer/autumn across most South African provinces while SINV cases did not show a clear seasonality and were detected in fewer South African provinces. MIDV could still be detected in blood samples via RT-PCR for up to 71,417 and 21 days after onset of signs in 4 horses respectively, suggesting prolonged replication relative to SINV which could only be detected in the initial sample. Phylogenetic analyses based on partial sequences of the nsP4 (MIDV n = 59 and SINV n = 7) and E1 (MIDV n = 45) genes, as well as full genome sequences (MIDV n = 6), clustered the MIDV and SINV strains from the present study with previously detected strains. MIDV infection appears to be more prevalent in horses than SINV infection based on RT-PCR results, however, prevalence estimates might be different when also considering serological surveillance data.
Keywords: Alphavirus; Middelburg virus; Sindbis virus; arbovirus; genome; horse; zoonotic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Zoonotic Alphaviruses in Fatal and Neurologic Infections in Wildlife and Nonequine Domestic Animals, South Africa.Emerg Infect Dis. 2020 Jun;26(6):1182-1191. doi: 10.3201/eid2606.191179. Emerg Infect Dis. 2020. PMID: 32441633 Free PMC article.
-
Sindbis and Middelburg Old World Alphaviruses Associated with Neurologic Disease in Horses, South Africa.Emerg Infect Dis. 2015 Dec;21(12):2225-9. doi: 10.3201/eid2112.150132. Emerg Infect Dis. 2015. PMID: 26583836 Free PMC article.
-
Detection of two alphaviruses: Middelburg virus and Sindbis virus from enzootic amplification cycles in southwestern Uganda.Front Microbiol. 2024 May 28;15:1394661. doi: 10.3389/fmicb.2024.1394661. eCollection 2024. Front Microbiol. 2024. PMID: 38863760 Free PMC article.
-
Sindbis virus as a human pathogen-epidemiology, clinical picture and pathogenesis.Rev Med Virol. 2016 Jul;26(4):221-41. doi: 10.1002/rmv.1876. Epub 2016 Mar 15. Rev Med Virol. 2016. PMID: 26990827 Review.
-
The Regulation of Translation in Alphavirus-Infected Cells.Viruses. 2018 Feb 8;10(2):70. doi: 10.3390/v10020070. Viruses. 2018. PMID: 29419763 Free PMC article. Review.
Cited by
-
A historical perspective on arboviruses of public health interest in Southern Africa.Pathog Glob Health. 2024 Mar;118(2):131-159. doi: 10.1080/20477724.2023.2290375. Epub 2023 Dec 11. Pathog Glob Health. 2024. PMID: 38082563 Free PMC article. Review.
-
Local Circulation of Sindbis Virus in Wild Birds and Horses, the Netherlands, 2021-2022.Emerg Infect Dis. 2025 Apr;31(4):863-866. doi: 10.3201/eid3104.241503. Emerg Infect Dis. 2025. PMID: 40133068 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

