Genomic and Epidemiological Features of COVID-19 in the Novosibirsk Region during the Beginning of the Pandemic
- PMID: 36146842
- PMCID: PMC9501018
- DOI: 10.3390/v14092036
Genomic and Epidemiological Features of COVID-19 in the Novosibirsk Region during the Beginning of the Pandemic
Abstract
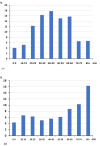
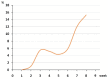
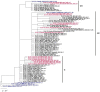
In this retrospective, single-center study, we conducted an analysis of 13,699 samples from different individuals obtained from the Federal Research Center of Fundamental and Translational Medicine, from 1 April to 30 May 2020 in Novosibirsk region (population 2.8 million people). We identified 6.49% positive for SARS-CoV-2 cases out of the total number of diagnostic tests, and 42% of them were from asymptomatic people. We also detected two asymptomatic people, who had no confirmed contact with patients with COVID-19. The highest percentage of positive samples was observed in the 80+ group (16.3%), while among the children and adults it did not exceed 8%. Among all the people tested, 2423 came from a total of 80 different destinations and only 27 of them were positive for SARS-CoV-2. Out of all the positive samples, 15 were taken for SARS-CoV-2 sequencing. According to the analysis of the genome sequences, the SARS-CoV-2 variants isolated in the Novosibirsk region at the beginning of the pandemic belonged to three phylogenetic lineages according to the Pangolin classification: B.1, B.1.1, and B.1.1.129. All Novosibirsk isolates contained the D614G substitution in the Spike protein, two isolates werecharacterized by an additional M153T mutation, and one isolate wascharacterized by the L5F mutation.
Keywords: COVID-19; Russia; SARS-CoV-2; epidemiology; first wave; phylogeny.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
SARS-CoV-2 genomic surveillance in Costa Rica: Evidence of a divergent population and an increased detection of a spike T1117I mutation.Infect Genet Evol. 2021 Aug;92:104872. doi: 10.1016/j.meegid.2021.104872. Epub 2021 Apr 24. Infect Genet Evol. 2021. PMID: 33905892 Free PMC article.
-
Large-scale sequencing of SARS-CoV-2 genomes from one region allows detailed epidemiology and enables local outbreak management.Microb Genom. 2021 Jun;7(6):000589. doi: 10.1099/mgen.0.000589. Microb Genom. 2021. PMID: 34184982 Free PMC article.
-
Molecular epidemiology of SARS-CoV-2 isolated from COVID-19 family clusters.BMC Med Genomics. 2021 Jun 1;14(1):144. doi: 10.1186/s12920-021-00990-3. BMC Med Genomics. 2021. PMID: 34074255 Free PMC article.
-
Evolutionary Tracking of SARS-CoV-2 Genetic Variants Highlights an Intricate Balance of Stabilizing and Destabilizing Mutations.mBio. 2021 Aug 31;12(4):e0118821. doi: 10.1128/mBio.01188-21. Epub 2021 Jul 20. mBio. 2021. PMID: 34281387 Free PMC article.
-
Phylogenetic and amino acid signature analysis of the SARS-CoV-2s lineages circulating in Tunisia.Infect Genet Evol. 2022 Aug;102:105300. doi: 10.1016/j.meegid.2022.105300. Epub 2022 May 10. Infect Genet Evol. 2022. PMID: 35552003 Free PMC article.
Cited by
-
The Development of the SARS-CoV-2 Epidemic in Different Regions of Siberia in the 2020-2022 Period.Viruses. 2023 Sep 27;15(10):2014. doi: 10.3390/v15102014. Viruses. 2023. PMID: 37896792 Free PMC article.
References
-
- Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., Blomberg W.R., Meigs D.D., Hasan M., Patel M., et al. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. - DOI - PMC - PubMed
-
- Van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

