A Novel Freshwater Cyanophage Mae-Yong1326-1 Infecting Bloom-Forming Cyanobacterium Microcystis aeruginosa
- PMID: 36146857
- PMCID: PMC9503304
- DOI: 10.3390/v14092051
A Novel Freshwater Cyanophage Mae-Yong1326-1 Infecting Bloom-Forming Cyanobacterium Microcystis aeruginosa
Abstract
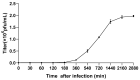
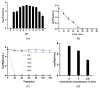
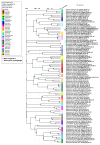
Microcystis aeruginosa is a major harmful cyanobacterium causing water bloom worldwide. Cyanophage has been proposed as a promising tool for cyanobacterial bloom. In this study, M. aeruginosa FACHB-1326 was used as an indicator host to isolate cyanophage from Lake Taihu. The isolated Microcystis cyanophage Mae-Yong1326-1 has an elliptical head of about 47 nm in diameter and a slender flexible tail of about 340 nm in length. Mae-Yong1326-1 could lyse cyanobacterial strains across three orders (Chroococcales, Nostocales, and Oscillatoriales) in the host range experiments. Mae-Yong1326-1 was stable in stability tests, maintaining high titers at 0-40 °C and at a wide pH range of 3-12. Mae-Yong 1326-1 has a burst size of 329 PFU/cell, which is much larger than the reported Microcystis cyanophages so far. The complete genome of Mae-Yong1326-1 is a double-stranded DNA of 48, 822 bp, with a G + C content of 71.80% and long direct terminal repeats (DTR) of 366 bp, containing 57 predicted ORFs. No Mae-Yong1326-1 ORF was found to be associated with virulence factor or antibiotic resistance. PASC scanning illustrated that the highest nucleotide sequence similarity between Mae-Yong1326-1 and all known phages in databases was only 17.75%, less than 70% (the threshold to define a genus), which indicates that Mae-Yong1326-1 belongs to an unknown new genus. In the proteomic tree based on genome-wide sequence similarities, Mae-Yong1326-1 distantly clusters with three unclassified Microcystis cyanophages (MinS1, Mwe-Yong1112-1, and Mwes-Yong2). These four Microcystis cyanophages form a monophyletic clade, which separates at a node from the other clade formed by two independent families (Zierdtviridae and Orlajensenviridae) of Caudoviricetes class. We propose to establish a new family to harbor the Microcystis cyanophages Mae-Yong1326-1, MinS1, Mwe-Yong1112-1, and Mwes-Yong2. This study enriched the understanding of freshwater cyanophages.
Keywords: Microcystis aeruginosa; cyanophage; genome; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Cyanophage Engineering for Algal Blooms Control.Viruses. 2024 Nov 6;16(11):1745. doi: 10.3390/v16111745. Viruses. 2024. PMID: 39599859 Free PMC article. Review.
-
A Novel Wide-Range Freshwater Cyanophage MinS1 Infecting the Harmful Cyanobacterium Microcystis aeruginosa.Viruses. 2022 Feb 20;14(2):433. doi: 10.3390/v14020433. Viruses. 2022. PMID: 35216026 Free PMC article.
-
A Novel Freshwater Cyanophage, Mae-Yong924-1, Reveals a New Family.Viruses. 2022 Jan 28;14(2):283. doi: 10.3390/v14020283. Viruses. 2022. PMID: 35215876 Free PMC article.
-
Genome sequence of the novel freshwater Microcystis cyanophage Mwe-Yong1112-1.Arch Virol. 2022 Nov;167(11):2371-2376. doi: 10.1007/s00705-022-05542-3. Epub 2022 Jul 20. Arch Virol. 2022. PMID: 35857150
-
Cyanophage infection in the bloom-forming cyanobacteria Microcystis aeruginosa in surface freshwater.Microbes Environ. 2012;27(4):350-5. doi: 10.1264/jsme2.me12037. Epub 2012 Oct 5. Microbes Environ. 2012. PMID: 23047146 Free PMC article. Review.
Cited by
-
Cyanophage Engineering for Algal Blooms Control.Viruses. 2024 Nov 6;16(11):1745. doi: 10.3390/v16111745. Viruses. 2024. PMID: 39599859 Free PMC article. Review.
-
Ecological Dynamics of Broad- and Narrow-Host-Range Viruses Infecting the Bloom-Forming Toxic Cyanobacterium Microcystis aeruginosa.Appl Environ Microbiol. 2023 Feb 28;89(2):e0211122. doi: 10.1128/aem.02111-22. Epub 2023 Jan 23. Appl Environ Microbiol. 2023. PMID: 36688685 Free PMC article.
References
-
- Whitton B.A., Potts M. Ecology of Cyanobacteria II. Springer, Dordrecht; Berlin/Heidelberg, Germany: 2012. Introduction to the cyanobacteria; pp. 1–13.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

