Use of LoopDeelab during the COVID-19 Pandemic: An Innovative Device for Field Diagnosis
- PMID: 36146869
- PMCID: PMC9505249
- DOI: 10.3390/v14092062
Use of LoopDeelab during the COVID-19 Pandemic: An Innovative Device for Field Diagnosis
Abstract
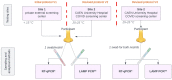
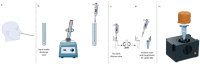
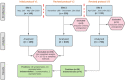
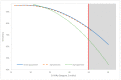
Rapid and accurate diagnosis of SARS-CoV-2 infection is essential for the management of the COVID-19 outbreak. RT-LAMP LoopDeetect COVID-19 (LoopDeescience, France) is a rapid molecular diagnostic tool which operates with the LoopDeelab (LoopDeescience, France) device. RAPID COVID is a prospective double-blind research protocol which was conducted to evaluate the concordance between Loopdeetect COVID-19 and RT-PCR Allplex 2019 n-Cov (Seegene, Korea). Between 11 May 2020 and 14 June 2021, a total of 1122 nasopharyngeal swab specimens were collected, of which 741 were finally analysed. There were 32 "positive" and "indeterminate" RT-PCR results. The intrinsic performances of Loopdeetect COVID-19 are equivalent to other commercial RT-LAMP PCR COVID-19 kits, with a sensitivity and specificity of 69.23% [CI 95%: 48.21-85.67] and 100% [CI 95%: 99.58-100.00], respectively. To the best of our knowledge, LoopDeelab is the only LAMP PCR diagnostic device allowing such a fast and reliable analysis with low-cost equipment; this makes it a new and innovative technology, designed for field use. This device being portable, the development of other detection kits will be useful for the management of epidemics with a high attack rate and would facilitate the rapid application of health measures.
Keywords: COVID-19; LAMP PCR; Loopdeelab; Loopdeetect; RAPID COVID; SARS-CoV-2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- World Health Organization (WHO) WHO Coronavirus (COVID-19) Dashboard. [(accessed on 23 June 2022)]. Available online: https://covid19.who.int/
-
- Lee S., Khoo V.S.L., Medriano C.A.D., Lee T., Park S.-Y., Bae S. Rapid and in-situ detection of fecal indicator bacteria in water using simple DNA extraction and portable loop mediated isothermal amplification (LAMP) PCR methods. Water Res. 2019;160:371–379. doi: 10.1016/j.watres.2019.05.049. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

