SNHG16 upregulation-induced positive feedback loop with YAP1/TEAD1 complex in Colorectal Cancer cell lines facilitates liver metastasis of colorectal cancer by modulating CTCs epithelial-mesenchymal transition
- PMID: 36147462
- PMCID: PMC9461660
- DOI: 10.7150/ijbs.73438
SNHG16 upregulation-induced positive feedback loop with YAP1/TEAD1 complex in Colorectal Cancer cell lines facilitates liver metastasis of colorectal cancer by modulating CTCs epithelial-mesenchymal transition
Abstract
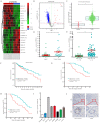
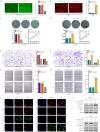
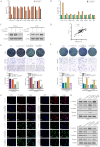
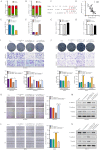
Circulating tumor cells (CTCs) are important precursors of colorectal cancer (CRC) metastasis. The epithelial-mesenchymal transition (EMT) process facilitates CTC invasion by allowing these cells to evade antimetastatic checkpoints to mediate distant metastasis. However, the specific molecular mechanism of tumor EMT remains largely unknown. Based on our previous research on the YAP1 pathway, we further studied the upstream molecule small nucleolar RNA host gene 16 (SNHG16), whose expression was correlated with advanced TNM stage, distant metastasis, and poor prognosis in CRC patients. Furthermore, loss- and gain-of-function assays revealed that SNHG16 promoted CRC colony formation, proliferation, migration, invasion, EMT, mesenchymal-like CTC generation, and liver metastasis through YAP1. Mechanistically, SNHG16 acted as a miRNA sponge to sequester miR-195-5p on Ago2, thereby protecting YAP1 from repression. Moreover, YAP1 bound TEA domain transcription factor 1 (TEAD1) to form a YAP1/TEAD1 complex, which in turn bound two sites in the promoter of SNHG16 and regulate SNHG16 transcription. Finally, in vivo experiments showed that the inhibition of SNHG16 suppressed tumor progression, and that YAP1 rescued the effect of SNHG16 on tumor progression. Herein, we have clarified a hitherto unexplored SNHG16-YAP1/TEAD1 positive feedback loop, that may be a candidate target for CRC treatment.
Keywords: CTCs; EMT; SNHG16; TEAD1; YAP1.
© The author(s).
Conflict of interest statement
Competing Interests: We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with this work.
Figures


References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: a cancer journal for clinicians. 2022;72:7–33. - PubMed
-
- Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM. et al. Cancer treatment and survivorship statistics, 2019. CA: a cancer journal for clinicians. 2019;69:363–85. - PubMed
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87–108. - PubMed
-
- van de Velde CJ, Boelens PG, Borras JM, Coebergh JW, Cervantes A, Blomqvist L. et al. EURECCA colorectal: multidisciplinary management: European consensus conference colon & rectum. Eur J Cancer. 2014;50:1. e- e34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

