Downregulation of PPARα mediates FABP1 expression, contributing to IgA nephropathy by stimulating ferroptosis in human mesangial cells
- PMID: 36147466
- PMCID: PMC9461665
- DOI: 10.7150/ijbs.74675
Downregulation of PPARα mediates FABP1 expression, contributing to IgA nephropathy by stimulating ferroptosis in human mesangial cells
Abstract
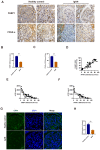
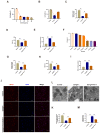
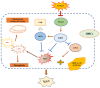
Immunoglobulin A nephropathy (IgAN) is the commonest primary glomerulonephritis, and a major cause of end-stage renal disease; however, its pathogenesis requires elucidation. Here, a hub gene, FABP1, and signaling pathway, PPARα, were selected as key in IgAN pathogenesis by combined weighted gene correlation network analysis of clinical traits and identification of differentially expressed genes from three datasets. FABP1 and PPARα levels were lower in IgAN than control kidney, and linearly positively correlated with one another, while FABP1 levels were negatively correlated with urinary albumin-to-creatinine ratio, and GPX4 levels were significantly decreased in IgAN. In human mesangial cells (HMCs), PPARα and FABP1 levels were significantly decreased after Gd-IgA1 stimulation and mitochondria appeared structurally damaged, while reactive oxygen species (ROS) and malondialdehyde (MDA) were significantly increased, and glutathione and GPX4 decreased, relative to controls. GPX4 levels were decreased, and those of ACSL4 increased on siPPARα and siFABP1 siRNA treatment. In PPARα lentivirus-transfected HMCs stimulated by Gd-IgA1, ROS, MDA, and ACSL4 were decreased; glutathione and GPX4, and immunofluorescence colocalization of PPARα and GPX4, increased; and damaged mitochondria reduced. Hence, PPARα pathway downregulation can reduce FABP1 expression, affecting GPX4 and ACSL4 levels, causing HMC ferroptosis, and contributing to IgAN pathogenesis.
Keywords: Fatty acid binding protein 1; Ferroptosis; Human mesangial cells; Immunoglobulin A nephropathy; Peroxisome proliferator-activated receptor α; Weighted gene correlation network analysis.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






Similar articles
-
Downregulation of CD36 alleviates IgA nephropathy by promoting autophagy and inhibiting extracellular matrix accumulation in mesangial cells.Int Immunopharmacol. 2025 Jan 10;144:113672. doi: 10.1016/j.intimp.2024.113672. Epub 2024 Nov 30. Int Immunopharmacol. 2025. PMID: 39616852
-
Sialylation of IgG inhibits the formation of galactose-deficient IgA1-containing immune complexes and protects mesangial cells from injury in IgA nephropathy.BMC Nephrol. 2022 Jan 11;23(1):25. doi: 10.1186/s12882-021-02657-8. BMC Nephrol. 2022. PMID: 35016642 Free PMC article.
-
Protective effect of phosphoenolpyruvate carboxykinase 1 on inflammation and fibrotic progression of IgA nephropathy.Ren Fail. 2025 Dec;47(1):2508297. doi: 10.1080/0886022X.2025.2508297. Epub 2025 May 29. Ren Fail. 2025. PMID: 40442892 Free PMC article.
-
Transferrin receptor engagement by polymeric IgA1 induces receptor expression and mesangial cell proliferation: role in IgA nephropathy.Contrib Nephrol. 2007;157:144-7. doi: 10.1159/000102457. Contrib Nephrol. 2007. PMID: 17495453 Review.
-
Molecular Insights into the Pathogenesis of IgA Nephropathy.Trends Mol Med. 2015 Dec;21(12):762-775. doi: 10.1016/j.molmed.2015.10.003. Epub 2015 Nov 21. Trends Mol Med. 2015. PMID: 26614735 Review.
Cited by
-
A Nutrient-Deficient Microenvironment Facilitates Ferroptosis Resistance via the FAM60A-PPAR Axis in Pancreatic Ductal Adenocarcinoma.Research (Wash D C). 2024 Feb 2;7:0300. doi: 10.34133/research.0300. eCollection 2024. Research (Wash D C). 2024. PMID: 38314086 Free PMC article.
-
Ferroptosis-Related Genes in IgA Nephropathy: Screening for Potential Targets of the Mechanism.Int J Genomics. 2024 Aug 14;2024:8851124. doi: 10.1155/2024/8851124. eCollection 2024. Int J Genomics. 2024. PMID: 39171207 Free PMC article.
-
Type I IFN in Glomerular Disease: Scarring beyond the STING.Int J Mol Sci. 2024 Feb 21;25(5):2497. doi: 10.3390/ijms25052497. Int J Mol Sci. 2024. PMID: 38473743 Free PMC article. Review.
-
Types of cell death and their relations to host immunological pathways.Aging (Albany NY). 2024 Aug 8;16(15):11755-11768. doi: 10.18632/aging.206035. Epub 2024 Aug 8. Aging (Albany NY). 2024. PMID: 39120579 Free PMC article. Review.
-
Ferroptosis is involved in passive Heymann nephritis in rats.Heliyon. 2023 Oct 17;9(10):e21050. doi: 10.1016/j.heliyon.2023.e21050. eCollection 2023 Oct. Heliyon. 2023. PMID: 37886789 Free PMC article.
References
-
- Chen A, Yang SS, Lin TJ. et al. IgA nephropathy: clearance kinetics of IgA-containing immune complexes. Seminars in immunopathology. 2018;40:539–43. - PubMed
-
- Manno C, Strippoli GF, D'Altri C. et al. A novel simpler histological classification for renal survival in IgA nephropathy: a retrospective study. American journal of kidney diseases: the official journal of the National Kidney Foundation. 2007;49:763–75. - PubMed
-
- Roberts IS. Pathology of IgA nephropathy. Nat Rev Nephrol. 2014;10:445–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

