Antibody interfaces revealed through structural mining
- PMID: 36147680
- PMCID: PMC9474289
- DOI: 10.1016/j.csbj.2022.08.048
Antibody interfaces revealed through structural mining
Abstract
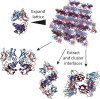
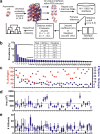
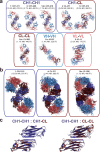
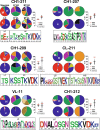
Antibodies are fundamental effectors of humoral immunity, and have become a highly successful class of therapeutics. There is increasing evidence that antibodies utilize transient homotypic interactions to enhance function, and elucidation of such interactions can provide insights into their biology and new opportunities for their optimization as drugs. Yet the transitory nature of weak interactions makes them difficult to investigate. Capitalizing on their rich structural data and high conservation, we have characterized all the ways that antibody fragment antigen-binding (Fab) regions interact crystallographically. This approach led to the discovery of previously unrealized interfaces between antibodies. While diverse interactions exist, β-sheet dimers and variable-constant elbow dimers are recurrent motifs. Disulfide engineering enabled interactions to be trapped and investigated structurally and functionally, providing experimental validation of the interfaces and illustrating their potential for optimization. This work provides first insight into previously undiscovered oligomeric interactions between antibodies, and enables new opportunities for their biotherapeutic optimization.
Keywords: Antibody; Cluster; Crystallography; Homotypic; Interface; Oligomer; Protein data bank; Structure.
© 2022 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Greg Lazar reports financial support was provided by Genentech USA Inc South San Francisco. Matthew G. Romei, Kannan Sankar, Kam Hon Hoi, Yanli Yang, Brandon Leonard, Gladys De Leon Boenig, Nikit Kumar, Marissa Matsumoto, Jian Payandeh, Seth F. Harris reports financial support was provided by Genentech USA Inc South San Francisco. Greg Lazar reports a relationship with Genentech USA Inc South San Francisco that includes: employment and equity or stocks. Matthew G. Romei, Kannan Sankar, Kam Hon Hoi, Yanli Yang, Brandon Leonard, Gladys De Leon Boenig, Nikit Kumar, Marissa Matsumoto, Jian Payandeh, Seth F. Harris reports a relationship with Genentech USA Inc South San Francisco that includes: employment and equity or stocks.
Figures


References
LinkOut - more resources
Full Text Sources
