Long non-coding RNA MALAT1 promotes Th2 differentiation by regulating microRNA-135b-5p/GATA-3 axis in children with allergic rhinitis
- PMID: 36149748
- PMCID: PMC11896592
- DOI: 10.1002/kjm2.12587
Long non-coding RNA MALAT1 promotes Th2 differentiation by regulating microRNA-135b-5p/GATA-3 axis in children with allergic rhinitis
Abstract
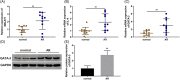
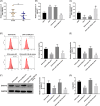
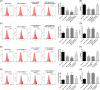
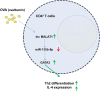
Allergic rhinitis (AR) threatens patient survival. CD4+ T cells play key roles in AR progression. Long non-coding RNAs (lncRNAs) are key regulators of cell differentiation. Therefore, we investigated the molecular mechanism of the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in AR. Expression levels of MALAT1, microRNA (miR)-135b-5p, interleukin-4 (IL-4), and GATA-binding protein 3 (GATA-3) in the nasal mucosa of AR patients were quantified. CD4+ T cells were isolated from the peripheral blood of healthy volunteers and treated with ovalbumin (OVA) and Th2 inducers. After MALAT1 and miR-135b-5p levels changed in CD4+ T cells, the proportion of IL-4-expressing cells and the levels of IL-4 and GATA-3 in OVA-induced CD4+ T cells were determined. Binding relationships among MALAT1, miR-135b-5p, and GATA-3 were predicted and verified. Rescue experiments were performed to confirm the role of the MALAT1/miR-135b-5p/GATA-3 axis in Th2 differentiation of CD4+ T cells. MALAT1, IL-4, and GATA-3 expression was upregulated, whereas miR-135b-5p expression was downregulated, in patients with AR. MALAT1 knockdown or miR-135b-5p overexpression in CD4+ T cells notably decreased the proportion of IL-4-expressing cells and downregulated GATA-3 and IL-4 expression in OVA-induced CD4+ T cells. MALAT1 and GATA-3 exhibited competitive binding toward miR-135b-5p. MALAT1 facilitated CD4+ T cell Th2 differentiation via the miR-135b-5p/GATA-3 axis. MALAT1 facilitated AR development by facilitating CD4+ T cell Th2 differentiation via the miR-135b-5p/GATA-3 axis. This study may provide guidance for clinical treatment of AR.
Keywords: GATA binding protein 3; T-helper 2 differentiation; allergic rhinitis; long non-coding RNA MALAT1; microRNA-135b-5p.
© 2022 The Authors. The Kaohsiung Journal of Medical Sciences published by John Wiley & Sons Australia, Ltd on behalf of Kaohsiung Medical University.
Conflict of interest statement
All authors declare no conflict of interest.
Figures

References
-
- Kalmarzi RN, Ataee P, Fathollahpour A, Behzadifar M, Moradi M, Sharifian F, et al. The prevalence of allergic rhinitis among Iranian children: a systematic review and meta‐analysis. Endocr Metab Immune Disord Drug Targets. 2020;20(2):189–97. - PubMed
-
- Testa D, Di Bari M, Nunziata M, Cristofaro G, Massaro G, Marcuccio G, et al. Allergic rhinitis and asthma assessment of risk factors in pediatric patients: a systematic review. Int J Pediatr Otorhinolaryngol. 2020;129:109759. - PubMed
-
- Kakli HA, Riley TD. Allergic rhinitis. Prim Care. 2016;43(3):465–75. - PubMed
-
- Ridolo E, Martignago I, Masieri S. Mechanisms of allergic diseases in otorhinolaryngology. J Biol Regul Homeost Agents. 2018;32(1 Suppl. 1):9–12. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

