Host-mycobiome metabolic interactions in health and disease
- PMID: 36151873
- PMCID: PMC9519009
- DOI: 10.1080/19490976.2022.2121576
Host-mycobiome metabolic interactions in health and disease
Abstract
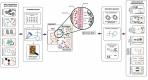
Fungal communities (mycobiome) have an important role in sustaining the resilience of complex microbial communities and maintenance of homeostasis. The mycobiome remains relatively unexplored compared to the bacteriome despite increasing evidence highlighting their contribution to host-microbiome interactions in health and disease. Despite being a small proportion of the total species, fungi constitute a large proportion of the biomass within the human microbiome and thus serve as a potential target for metabolic reprogramming in pathogenesis and disease mechanism. Metabolites produced by fungi shape host niches, induce immune tolerance and changes in their levels prelude changes associated with metabolic diseases and cancer. Given the complexity of microbial interactions, studying the metabolic interplay of the mycobiome with both host and microbiome is a demanding but crucial task. However, genome-scale modelling and synthetic biology can provide an integrative platform that allows elucidation of the multifaceted interactions between mycobiome, microbiome and host. The inferences gained from understanding mycobiome interplay with other organisms can delineate the key role of the mycobiome in pathophysiology and reveal its role in human disease.
Keywords: Mycobiome; host-mycobiome interaction; metabolism; microbiome; secondary metabolism; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
