Stress-triggered hematopoietic stem cell proliferation relies on PrimPol-mediated repriming
- PMID: 36152632
- PMCID: PMC10251193
- DOI: 10.1016/j.molcel.2022.09.009
Stress-triggered hematopoietic stem cell proliferation relies on PrimPol-mediated repriming
Abstract
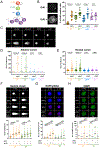
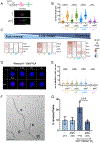
Stem cell division is linked to tumorigenesis by yet-elusive mechanisms. The hematopoietic system reacts to stress by triggering hematopoietic stem and progenitor cell (HSPC) proliferation, which can be accompanied by chromosomal breakage in activated hematopoietic stem cells (HSCs). However, whether these lesions persist in their downstream progeny and induce a canonical DNA damage response (DDR) remains unclear. Inducing HSPC proliferation by simulated viral infection, we report that the associated DNA damage is restricted to HSCs and that proliferating HSCs rewire their DDR upon endogenous and clastogen-induced damage. Combining transcriptomics, single-cell and single-molecule assays on murine bone marrow cells, we found accelerated fork progression in stimulated HSPCs, reflecting engagement of PrimPol-dependent repriming, at the expense of replication fork reversal. Ultimately, competitive bone marrow transplantation revealed the requirement of PrimPol for efficient HSC amplification and bone marrow reconstitution. Hence, fine-tuning replication fork plasticity is essential to support stem cell functionality upon proliferation stimuli.
Keywords: DNA damage response; DNA replication; Primpol; bone marrow reconstitution; hematopoietic stem cells; induced proliferation; replication fork plasticity; repriming; stem cell division.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

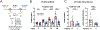
Similar articles
-
DNA damage tolerance in hematopoietic stem and progenitor cells in mice.Proc Natl Acad Sci U S A. 2017 Aug 15;114(33):E6875-E6883. doi: 10.1073/pnas.1706508114. Epub 2017 Jul 31. Proc Natl Acad Sci U S A. 2017. PMID: 28761001 Free PMC article.
-
DNA fork remodeling proteins, Zranb3 and Smarcal1, are uniquely essential for aging hematopoiesis.Aging Cell. 2024 Nov;23(11):e14281. doi: 10.1111/acel.14281. Epub 2024 Jul 23. Aging Cell. 2024. PMID: 39044358 Free PMC article.
-
Type I IFNs drive hematopoietic stem and progenitor cell collapse via impaired proliferation and increased RIPK1-dependent cell death during shock-like ehrlichial infection.PLoS Pathog. 2018 Aug 6;14(8):e1007234. doi: 10.1371/journal.ppat.1007234. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30080899 Free PMC article.
-
Tolerating DNA damage by repriming: Gap filling in the spotlight.DNA Repair (Amst). 2024 Oct;142:103758. doi: 10.1016/j.dnarep.2024.103758. Epub 2024 Aug 30. DNA Repair (Amst). 2024. PMID: 39236419 Review.
-
PRIMPOL ready, set, reprime!Crit Rev Biochem Mol Biol. 2021 Feb;56(1):17-30. doi: 10.1080/10409238.2020.1841089. Epub 2020 Nov 12. Crit Rev Biochem Mol Biol. 2021. PMID: 33179522 Free PMC article. Review.
Cited by
-
Comprehensive interrogation of synthetic lethality in the DNA damage response.Nature. 2025 Apr;640(8060):1093-1102. doi: 10.1038/s41586-025-08815-4. Epub 2025 Apr 9. Nature. 2025. PMID: 40205037 Free PMC article.
-
Nuclear actin polymerization rapidly mediates replication fork remodeling upon stress by limiting PrimPol activity.Nat Commun. 2023 Nov 28;14(1):7819. doi: 10.1038/s41467-023-43183-5. Nat Commun. 2023. PMID: 38016948 Free PMC article.
-
Proteogenetic drug response profiling elucidates targetable vulnerabilities of myelofibrosis.Nat Commun. 2023 Oct 12;14(1):6414. doi: 10.1038/s41467-023-42101-z. Nat Commun. 2023. PMID: 37828014 Free PMC article.
-
RAD51 restricts DNA over-replication from re-activated origins.EMBO J. 2024 Mar;43(6):1043-1064. doi: 10.1038/s44318-024-00038-z. Epub 2024 Feb 15. EMBO J. 2024. PMID: 38360996 Free PMC article.
-
Polymerase iota (Pol ι) prevents PrimPol-mediated nascent DNA synthesis and chromosome instability.Sci Adv. 2023 Apr 14;9(15):eade7997. doi: 10.1126/sciadv.ade7997. Epub 2023 Apr 14. Sci Adv. 2023. PMID: 37058556 Free PMC article.
References
-
- Adolfsson J, Borge OJ, Bryder D, Theilgaard-Mönch K, Åstrand-Grundström I, Sitnicka E, Sasaki Y, and Jacobsen SEW (2001). Upregulation of Flt3 Expression within the Bone Marrow Lin−Scal+c-kit+ Stem Cell Compartment Is Accompanied by Loss of Self-Renewal Capacity. Immunity 15, 659–669. 10.1016/s1074-7613(01)00220-5. - DOI - PubMed
-
- Ahuja AK, Jodkowska K, Teloni F, Bizard AH, Zellweger R, Herrador R, Ortega S, Hickson ID, Altmeyer M, Mendez J, et al. (2016). A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nature Communications 7, 10660. 10.1038/ncomms10660. - DOI - PMC - PubMed
-
- Bai G, Kermi C, Stoy H, Schiltz CJ, Bacal J, Zaino AM, Hadden MK, Eichman BF, Lopes M, and Cimprich KA (2020). HLTF Promotes Fork Reversal, Limiting Replication Stress Resistance and Preventing Multiple Mechanisms of Unrestrained DNA Synthesis. Mol Cell 10.1016/j.molcel.2020.04.031. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

