RNase HI Depletion Strongly Potentiates Cell Killing by Rifampicin in Mycobacteria
- PMID: 36154174
- PMCID: PMC9578417
- DOI: 10.1128/aac.02091-21
RNase HI Depletion Strongly Potentiates Cell Killing by Rifampicin in Mycobacteria
Abstract
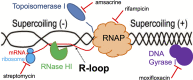
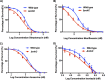
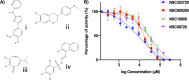
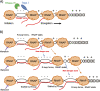
Multidrug-resistant (MDR) tuberculosis (TB) is defined by the resistance of Mycobacterium tuberculosis, the causative organism, to the first-line antibiotics rifampicin and isoniazid. Mitigating or reversing resistance to these drugs offers a means of preserving and extending their use in TB treatment. R-loops are RNA/DNA hybrids that are formed in the genome during transcription, and they can be lethal to the cell if not resolved. RNase HI is an enzyme that removes R-loops, and this activity is essential in M. tuberculosis: knockouts of rnhC, the gene encoding RNase HI, are nonviable. This essentiality makes it a candidate target for the development of new antibiotics. In the model organism Mycolicibacterium smegmatis, RNase HI activity is provided by two enzymes, RnhA and RnhC. We show that the partial depletion of RNase HI activity in M. smegmatis, by knocking out either of the genes encoding RnhA or RnhC, led to the accumulation of R-loops. The sensitivity of the knockout strains to the antibiotics moxifloxacin, streptomycin, and rifampicin was increased, the latter by a striking near 100-fold. We also show that R-loop accumulation accompanies partial transcriptional inhibition, suggesting a mechanistic basis for the synergy between RNase HI depletion and rifampicin. A model of how transcriptional inhibition can potentiate R-loop accumulation is presented. Finally, we identified four small molecules that inhibit recombinant RnhC activity and that also potentiated rifampicin activity in whole-cell assays against M. tuberculosis, supporting an on-target mode of action and providing the first step in developing a new class of antimycobacterial drug.
Keywords: R-loop; RNase HI; antibiotic development; antibiotic resistance; antibiotic synergy; rifampicin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The C-Terminal Acid Phosphatase Module of the RNase HI Enzyme RnhC Controls Rifampin Sensitivity and Light-Dependent Colony Pigmentation of Mycobacterium smegmatis.J Bacteriol. 2023 Apr 25;205(4):e0043122. doi: 10.1128/jb.00431-22. Epub 2023 Mar 14. J Bacteriol. 2023. PMID: 36916909 Free PMC article.
-
Occurrence of disputed rpoB mutations among Mycobacterium tuberculosis isolates phenotypically susceptible to rifampicin in a country with a low incidence of multidrug-resistant tuberculosis.BMC Infect Dis. 2019 Jan 3;19(1):3. doi: 10.1186/s12879-018-3638-z. BMC Infect Dis. 2019. PMID: 30606116 Free PMC article.
-
Division of labor among Mycobacterium smegmatis RNase H enzymes: RNase H1 activity of RnhA or RnhC is essential for growth whereas RnhB and RnhA guard against killing by hydrogen peroxide in stationary phase.Nucleic Acids Res. 2017 Jan 9;45(1):1-14. doi: 10.1093/nar/gkw1046. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899559 Free PMC article.
-
Molecular genetics of drug resistance in Mycobacterium tuberculosis.J Antimicrob Chemother. 1994 Sep;34(3):313-9. doi: 10.1093/jac/34.3.313. J Antimicrob Chemother. 1994. PMID: 7829406 Review.
-
[Molecular mechanisms of multidrug resistance in Mycobacterium tuberculosis].J UOEH. 2000 Sep 1;22(3):269-82. doi: 10.7888/juoeh.22.269. J UOEH. 2000. PMID: 11019393 Review. Japanese.
Cited by
-
Identification of gene targets that potentiate the action of rifampicin on Mycobacterium bovis BCG.Microbiology (Reading). 2024 Aug;170(8):001488. doi: 10.1099/mic.0.001488. Microbiology (Reading). 2024. PMID: 39150447 Free PMC article.
-
The C-Terminal Acid Phosphatase Module of the RNase HI Enzyme RnhC Controls Rifampin Sensitivity and Light-Dependent Colony Pigmentation of Mycobacterium smegmatis.J Bacteriol. 2023 Apr 25;205(4):e0043122. doi: 10.1128/jb.00431-22. Epub 2023 Mar 14. J Bacteriol. 2023. PMID: 36916909 Free PMC article.
-
Assessment for antibiotic resistance in Helicobacter pylori: A practical and interpretable machine learning model based on genome-wide genetic variation.Virulence. 2025 Dec;16(1):2481503. doi: 10.1080/21505594.2025.2481503. Epub 2025 Mar 21. Virulence. 2025. PMID: 40119500 Free PMC article.
-
New option: targeting RNase J and RNase HI in the fight against multi-drug-resistant tuberculosis.Ann Med Surg (Lond). 2024 Mar 4;86(5):2376-2378. doi: 10.1097/MS9.0000000000001859. eCollection 2024 May. Ann Med Surg (Lond). 2024. PMID: 38694338 Free PMC article. No abstract available.
References
-
- O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Wellcome Trust, London, UK.
-
- World Health Organisation. 2020. Consolidated guidelines on tuberculosis. https://www.who.int/publications/i/item/9789240007048.
-
- Conradie F, Diacon AH, Ngubane N, Howell P, Everitt D, Crook AM, Mendel CM, Egizi E, Moreira J, Timm J, McHugh TD, Wills GH, Bateson A, Hunt R, Van Niekerk C, Li M, Olugbosi M, Spigelman M, Nix TBTT, Nix-TB Trial Team. 2020. Treatment of highly drug-resistant pulmonary tuberculosis. N Engl J Med 382:893–902. 10.1056/NEJMoa1901814. - DOI - PMC - PubMed
-
- Drolet M, Phoenix P, Menzel R, Massé E, Liu LF, Crouch RJ. 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli ΔtopA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA 92:3526–3530. 10.1073/pnas.92.8.3526. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

