Microbial Interdomain Interactions Delineate the Disruptive Intestinal Homeostasis in Clostridioides difficile Infection
- PMID: 36154277
- PMCID: PMC9602525
- DOI: 10.1128/spectrum.00502-22
Microbial Interdomain Interactions Delineate the Disruptive Intestinal Homeostasis in Clostridioides difficile Infection
Abstract
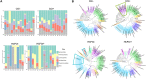
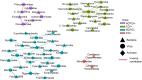
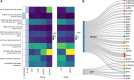
Clostridioides difficile infection (CDI) creates an imbalance in the intestinal microbiota due to the interaction of the components making up this ecosystem, but little is known about the impact of this disease on other microbial members. This work has thus been aimed at evaluating the taxonomic composition, potential gene-associated functions, virulence factors, and antimicrobial resistance profiles of gut microbiomes. A total of 48 DNA samples obtained from patients with health care facility-acquired (HCFO) and community-onset (CO) diarrhea were distributed in the following four groups according to CDI status: HCFO/+ (n = 13), HCFO/- (n = 8), CO/+ (n = 13), and CO/- (n = 14). These samples were subjected to shotgun metagenomics sequencing. Although the CDI groups' microbiota had microbiome alterations, the greatest imbalance was observed in the in the HCFO+/- groups, with an increase in common pathogens and phage populations, as well as a decrease in beneficial microorganisms that leads to a negative impact on some intestinal homeostasis-related metabolic processes. A reduction in the relative abundance of butyrate metabolism-associated genes was also detected in the HCFO groups (P < 0.01), with an increase in some virulence factors and antibiotic-resistance markers. A set of 51 differentially abundant species in the groups with potential association to CDI enabled its characterization, leading to their spatial separation by onset. Strong correlations between phages and some archaeal and bacterial phyla were identified. This highlighted the need to study the microbiota's various components since their imbalance is multifactorial, with some pathogens contributing to a greater or lesser extent because of their interaction with the ecosystem they inhabit. IMPORTANCE Clostridioides difficile infection represents a serious public health problem in different countries due to its high morbi-mortality and the high costs it represents for health care systems. Studies have shown the impact of this infection on intestinal microbiome homeostasis, mainly on bacterial populations. Our research provides evidence of the impact of CDI at both the compositional (bacteria, archaea, and viruses), and functional levels, allowing us to understand that the alterations of the microbiota occur systemically and are caused by multiple perturbations generated by different members of the microbiota as well as by some pathogens that take advantage of the imbalance to proliferate. Likewise, the 51 differentially abundant species in the study groups with potential association to CDI found in this study could help us envisage future treatments against this and other inflammatory diseases, improving future therapeutic options for patients.
Keywords: Clostridioides difficile; biomarkers; gut microbiome; interdomain interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Magill SS, O’Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle J, Wilson LE, Kainer MA, Lynfield R, Greissman S, Ray SM, Beldavs Z, Gross C, Bamberg W, Sievers M, Concannon C, Buhr N, Warnke L, Maloney M, Ocampo V, Brooks J, Oyewumi T, Sharmin S, Richards K, Rainbow J, Samper M, Hancock EB, Leaptrot D, Scalise E, Badrun F, Phelps R, Edwards JR. 2018. Changes in prevalence of health care-associated infections in U.S. hospitals. N Engl J Med 379:1732–1744. doi:10.1056/NEJMoa1801550. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

