GITR Ligation Improves Anti-PD1-Mediated Restoration of Human MMR-Proficient Colorectal Carcinoma Tumor-Derived T Cells
- PMID: 36155259
- PMCID: PMC9672455
- DOI: 10.1016/j.jcmgh.2022.09.007
GITR Ligation Improves Anti-PD1-Mediated Restoration of Human MMR-Proficient Colorectal Carcinoma Tumor-Derived T Cells
Abstract
Background & aims: In contrast to mismatch repair deficient colorectal carcinoma (CRC), MMR proficient (pMMR) CRC does not respond to immune checkpoint blockade. We studied immune checkpoint stimulation via glucocorticoid-induced tumor necrosis factor receptor-related protein (GITR) on ex vivo functionality of human tumor-infiltrating lymphocytes (TIL) isolated from pMMR primary CRC and liver metastases (CRLM).
Methods: Using lymphocytes from resected tumor, adjacent tissues, and peripheral blood mononuclear cells (PBMC) of 132 pMMR primary CRC or CRLM patients, we determined GITR expression and the in vitro T-cell agonistic activity of recombinant GITR ligation.
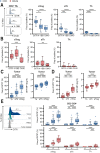
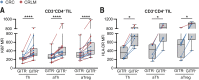
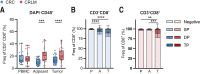
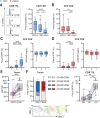
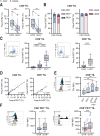
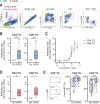
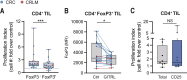
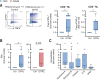
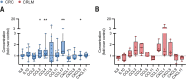
Results: Here, we show that GITR was overexpressed on TIL when compared with other stimulatory immune checkpoints (4-1BB, OX40). Its expression was enhanced in TIL compared with PBMC and adjacent tissues. Among CD4+ TIL, GITR expression was primarily expressed by CD45RA- FoxP3hi activated regulatory T cells. Within CD8+ TIL, GITR was predominantly expressed on functionally exhausted and putative tumor-reactive CD103+ CD39+ TIL. Strikingly, recombinant GITRL reinvigorated ex vivo TIL responses by significantly enhancing CD4+ and CD8+ TIL numbers. Dual treatment with GITRL and nivolumab (anti-PD1) enhanced CD8+ TIL expansion compared with GITRL monotherapy. Moreover, GITRL/anti-PD1 dual therapy further improved anti-PD1-mediated reinvigoration of interferon gamma secretion by exhausted CD8 TIL from primary CRC.
Conclusions: GITR is overexpressed on CD4+ and CD8+ TIL from pMMR CRC and CRLM. Agonistic targeting of GITR enhances ex vivo human TIL functionality and may therefore be a promising approach for novel monotherapy or combined immunotherapies in primary pMRR CRC and CRLM.
Keywords: Colorectal carcinoma; Immune Checkpoint Stimulation; Liver metastasis; Microsatellite Stable; TNF Receptor Superfamily; Tumor-Infiltrating Lymphocytes.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- IARC Fact sheets colorectal cancer by Globocan. 2020. 2020. https://gco.iarc.fr/today/data/factsheets/cancers/10_8_9-Colorectum-fact... Available at:
-
- Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394:1467–1480. - PubMed
-
- Guinney J., Dienstmann R., Wang X., De Reyniès A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., Bot B.M., Morris J.S., Simon I.M., Gerster S., Fessler E., De Sousa E Melo F., Missiaglia E., Ramay H., Barras D., Homicsko K., Maru D., Manyam G.C., Broom B., Boige V., Perez-Villamil B., Laeras T., Salazar R., Gray J.W., Hanahan D., Tabernero J., Berards R., Friend S.H., Laurent-Puig P., Medema J.P., Sadanandam A., Wessels L., Delorenzi M., Kopetz S., Vermeulen L., Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. - PMC - PubMed
-
- Combes A.J., Samad B., Tsui J., Chew N.W., Yan P., Reeder G.C., Kushnoor D., Shen A., Davidson B., Barczak A.J., Adkisson M., Edwards A., Naser M., Barry K.C., Courau T., Hammoudi T., Argüello R.J., Rao A.A., Olshen A.B., Immunoprolifer Consortium. Cai C., Zhan J., Daves K.C., Kelley R.K., Chapman J.S., Atreya C.L., Patel A., Daud A., Ha P., Diaz A.A., Krat J.R., Collisson E.A., Fragiadakis G.K., Erle D.J., Boissonnas A., Asthana S., Chan V., Krummel M.F. Discovering dominant tumor immune archetypes in a pan-cancer census. Cell. 2022;185:184–203. - PMC - PubMed
-
- Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoué F., Bruneval P., Cugnenc P., Trajanoski Z., Fridman W., Pagès F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

