Post-phagocytosis activation of NLRP3 inflammasome by two novel T6SS effectors
- PMID: 36155655
- PMCID: PMC9545535
- DOI: 10.7554/eLife.82766
Post-phagocytosis activation of NLRP3 inflammasome by two novel T6SS effectors
Abstract
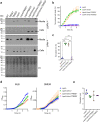
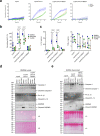
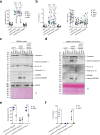
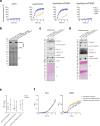
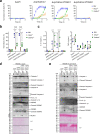
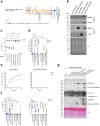
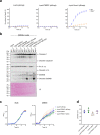
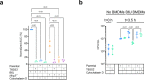
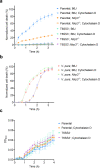
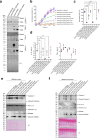
The type VI secretion system (T6SS) is used by bacteria to deliver toxic effectors directly into target cells. Most T6SSs mediate antibacterial activities, whereas the potential anti-eukaryotic role of T6SS remains understudied. Here, we found a Vibrio T6SS that delivers two novel effectors into mammalian host immune cells. We showed that these effectors induce a pyroptotic cell death in a phagocytosis-dependent manner; we identified the NLRP3 inflammasome as being the underlying mechanism leading to the T6SS-induced pyroptosis. Moreover, we identified a compensatory T6SS-induced pathway that is activated upon inhibition of the canonical pyroptosis pathway. Genetic analyses revealed possible horizontal spread of this T6SS and its anti-eukaryotic effectors into emerging pathogens in the marine environment. Our findings reveal novel T6SS effectors that activate the host inflammasome and possibly contribute to virulence and to the emergence of bacterial pathogens.
Keywords: T6SS; caspase; cell death; gasdermin; immunology; infectious disease; inflammation; microbiology; mouse; pyroptosis; secretion system; vibrio proteolyticus.
© 2022, Cohen et al.
Conflict of interest statement
HC, NB, CF, LE, DS, MG No competing interests declared
Figures


Similar articles
-
The Bacterial T6SS Effector EvpP Prevents NLRP3 Inflammasome Activation by Inhibiting the Ca2+-Dependent MAPK-Jnk Pathway.Cell Host Microbe. 2017 Jan 11;21(1):47-58. doi: 10.1016/j.chom.2016.12.004. Cell Host Microbe. 2017. PMID: 28081443
-
Multiple T6SSs, Mobile Auxiliary Modules, and Effectors Revealed in a Systematic Analysis of the Vibrio parahaemolyticus Pan-Genome.mSystems. 2022 Dec 20;7(6):e0072322. doi: 10.1128/msystems.00723-22. Epub 2022 Oct 13. mSystems. 2022. PMID: 36226968 Free PMC article.
-
The Antibacterial and Anti-Eukaryotic Type VI Secretion System MIX-Effector Repertoire in Vibrionaceae.Mar Drugs. 2018 Nov 4;16(11):433. doi: 10.3390/md16110433. Mar Drugs. 2018. PMID: 30400344 Free PMC article.
-
Does Pyroptosis Play a Role in Inflammasome-Related Disorders?Int J Mol Sci. 2022 Sep 9;23(18):10453. doi: 10.3390/ijms231810453. Int J Mol Sci. 2022. PMID: 36142364 Free PMC article. Review.
-
Bacterial type VI secretion system (T6SS): an evolved molecular weapon with diverse functionality.Biotechnol Lett. 2023 Mar;45(3):309-331. doi: 10.1007/s10529-023-03354-2. Epub 2023 Jan 23. Biotechnol Lett. 2023. PMID: 36683130 Review.
Cited by
-
The multiple roles of the NlpC_P60 peptidase family in mycobacteria - an underexplored target for antimicrobial drug discovery.FEBS Lett. 2025 May;599(9):1203-1221. doi: 10.1002/1873-3468.70021. Epub 2025 Mar 3. FEBS Lett. 2025. PMID: 40028658 Free PMC article. Review.
-
Strategies of bacterial detection by inflammasomes.Cell Chem Biol. 2024 May 16;31(5):835-850. doi: 10.1016/j.chembiol.2024.03.009. Epub 2024 Apr 17. Cell Chem Biol. 2024. PMID: 38636521 Free PMC article. Review.
-
Function and Global Regulation of Type III Secretion System and Flagella in Entomopathogenic Nematode Symbiotic Bacteria.Int J Mol Sci. 2024 Jul 10;25(14):7579. doi: 10.3390/ijms25147579. Int J Mol Sci. 2024. PMID: 39062822 Free PMC article.
-
Vibrio parahaemolyticus T6SS2 effector repertoires.Gut Microbes. 2023 Jan-Dec;15(1):2178795. doi: 10.1080/19490976.2023.2178795. Gut Microbes. 2023. PMID: 36803660 Free PMC article.
-
The RIX domain defines a class of polymorphic T6SS effectors and secreted adaptors.Nat Commun. 2023 Aug 17;14(1):4983. doi: 10.1038/s41467-023-40659-2. Nat Commun. 2023. PMID: 37591831 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

