Mouse models of COVID-19 recapitulate inflammatory pathways rather than gene expression
- PMID: 36155667
- PMCID: PMC9536645
- DOI: 10.1371/journal.ppat.1010867
Mouse models of COVID-19 recapitulate inflammatory pathways rather than gene expression
Abstract
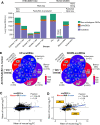
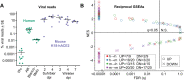
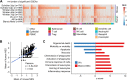
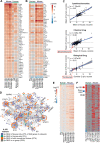
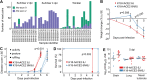
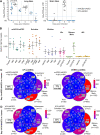
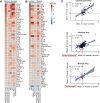
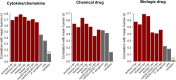
How well mouse models recapitulate the transcriptional profiles seen in humans remains debatable, with both conservation and diversity identified in various settings. Herein we use RNA-Seq data and bioinformatics approaches to analyze the transcriptional responses in SARS-CoV-2 infected lungs, comparing 4 human studies with the widely used K18-hACE2 mouse model, a model where hACE2 is expressed from the mouse ACE2 promoter, and a model that uses a mouse adapted virus and wild-type mice. Overlap of single copy orthologue differentially expressed genes (scoDEGs) between human and mouse studies was generally poor (≈15-35%). Rather than being associated with batch, sample treatment, viral load, lung damage or mouse model, the poor overlaps were primarily due to scoDEG expression differences between species. Importantly, analyses of immune signatures and inflammatory pathways illustrated highly significant concordances between species. As immunity and immunopathology are the focus of most studies, these mouse models can thus be viewed as representative and relevant models of COVID-19.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
SARS-CoV-2 Causes Lung Infection without Severe Disease in Human ACE2 Knock-In Mice.J Virol. 2022 Jan 12;96(1):e0151121. doi: 10.1128/JVI.01511-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668780 Free PMC article.
-
Lung Expression of Human Angiotensin-Converting Enzyme 2 Sensitizes the Mouse to SARS-CoV-2 Infection.Am J Respir Cell Mol Biol. 2021 Jan;64(1):79-88. doi: 10.1165/rcmb.2020-0354OC. Am J Respir Cell Mol Biol. 2021. PMID: 32991819 Free PMC article.
-
The K18-Human ACE2 Transgenic Mouse Model Recapitulates Non-severe and Severe COVID-19 in Response to an Infectious Dose of the SARS-CoV-2 Virus.J Virol. 2022 Jan 12;96(1):e0096421. doi: 10.1128/JVI.00964-21. Epub 2021 Oct 20. J Virol. 2022. PMID: 34668775 Free PMC article.
-
Infectious Clones Produce SARS-CoV-2 That Causes Severe Pulmonary Disease in Infected K18-Human ACE2 Mice.mBio. 2021 Apr 20;12(2):e00819-21. doi: 10.1128/mBio.00819-21. mBio. 2021. PMID: 33879586 Free PMC article.
-
K18- and CAG-hACE2 Transgenic Mouse Models and SARS-CoV-2: Implications for Neurodegeneration Research.Molecules. 2022 Jun 28;27(13):4142. doi: 10.3390/molecules27134142. Molecules. 2022. PMID: 35807384 Free PMC article. Review.
Cited by
-
The effects of iron deficient and high iron diets on SARS-CoV-2 lung infection and disease.Front Microbiol. 2024 Sep 4;15:1441495. doi: 10.3389/fmicb.2024.1441495. eCollection 2024. Front Microbiol. 2024. PMID: 39296289 Free PMC article.
-
Monoclonal Antibodies Specific for SARS-CoV-2 Spike Protein Suitable for Multiple Applications for Current Variants of Concern.Viruses. 2022 Dec 31;15(1):139. doi: 10.3390/v15010139. Viruses. 2022. PMID: 36680179 Free PMC article.
-
CCR5/CXCR3 antagonist TAK-779 prevents diffuse alveolar damage of the lung in the murine model of the acute respiratory distress syndrome.Front Pharmacol. 2024 Feb 21;15:1351655. doi: 10.3389/fphar.2024.1351655. eCollection 2024. Front Pharmacol. 2024. PMID: 38449806 Free PMC article.
-
A highly susceptible hACE2-transgenic mouse model for SARS-CoV-2 research.Front Microbiol. 2024 Feb 7;15:1348405. doi: 10.3389/fmicb.2024.1348405. eCollection 2024. Front Microbiol. 2024. PMID: 38389533 Free PMC article.
-
Tracking inflammation resolution signatures in lungs after SARS-CoV-2 omicron BA.1 infection of K18-hACE2 mice.PLoS One. 2024 Nov 12;19(11):e0302344. doi: 10.1371/journal.pone.0302344. eCollection 2024. PLoS One. 2024. PMID: 39531435 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

