PRC2-independent actions of H3.3K27M in embryonic stem cell differentiation
- PMID: 36156096
- PMCID: PMC9976889
- DOI: 10.1093/nar/gkac800
PRC2-independent actions of H3.3K27M in embryonic stem cell differentiation
Erratum in
-
Correction to 'PRC2-independent actions of H3.3K27M in embryonic stem cell differentiation'.Nucleic Acids Res. 2023 Feb 28;51(4):1996. doi: 10.1093/nar/gkad018. Nucleic Acids Res. 2023. PMID: 36625287 Free PMC article. No abstract available.
Abstract
The histone H3 variant, H3.3, is localized at specific regions in the genome, especially promoters and active enhancers, and has been shown to play important roles in development. A lysine to methionine substitution in position 27 (H3.3K27M) is a main cause of Diffuse Intrinsic Pontine Glioma (specifically Diffuse Midline Glioma, K27M-mutant), a lethal type of pediatric cancer. H3.3K27M has a dominant-negative effect by inhibiting the Polycomb Repressor Complex 2 (PRC2) activity. Here, we studied the immediate, genome-wide, consequences of the H3.3K27M mutation independent of PRC2 activity. We developed Doxycycline (Dox)-inducible mouse embryonic stem cells (ESCs) carrying a single extra copy of WT-H3.3, H3.3K27M and H3.3K27L, all fused to HA. We performed RNA-Seq and ChIP-Seq at different times following Dox induction in undifferentiated and differentiated ESCs. We find increased binding of H3.3 around transcription start sites in cells expressing both H3.3K27M and H3.3K27L compared with WT, but not in cells treated with PRC2 inhibitors. Differentiated cells carrying either H3.3K27M or H3.3K27L retain expression of ESC-active genes, in expense of expression of genes related to neuronal differentiation. Taken together, our data suggest that a modifiable H3.3K27 is required for proper histone incorporation and cellular maturation, independent of PRC2 activity.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
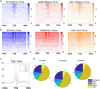



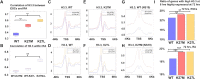
References
-
- Allis C.D., Jenuwein T.. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016; 17:487–500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

