The yeast 2-micron plasmid Rep2 protein has Rep1-independent partitioning function
- PMID: 36156142
- PMCID: PMC9561267
- DOI: 10.1093/nar/gkac810
The yeast 2-micron plasmid Rep2 protein has Rep1-independent partitioning function
Abstract
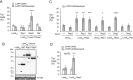
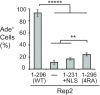
Equal partitioning of the multi-copy 2-micron plasmid of the budding yeast Saccharomyces cerevisiae requires association of the plasmid Rep1 and Rep2 proteins with the plasmid STB partitioning locus. Determining how the Rep proteins contribute has been complicated by interactions between the components. Here, each Rep protein was expressed fused to the DNA-binding domain of the bacterial repressor protein LexA in yeast harboring a replication-competent plasmid that had LexA-binding sites but lacked STB. Plasmid transmission to daughter cells was increased only by Rep2 fusion expression. Neither Rep1 nor a functional RSC2 complex (a chromatin remodeler required for 2-micron plasmid partitioning) were needed for the improvement. Deletion analysis showed the carboxy-terminal 65 residues of Rep2 were required and sufficient for this Rep1-independent inheritance. Mutation of a conserved basic motif in this domain impaired Rep1-independent and Rep protein/STB-dependent plasmid partitioning. Our findings suggest Rep2, which requires Rep1 and the RSC2 complex for functional association with STB, directly participates in 2-micron plasmid partitioning by linking the plasmid to a host component that is efficiently partitioned during cell division. Further investigation is needed to reveal the host factor targeted by Rep2 that contributes to the survival of these plasmids in their budding yeast hosts.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Murray A.W., Szostak J.W.. Pedigree analysis of plasmid segregation in yeast. Cell. 1983; 34:961–970. - PubMed
-
- Gehlen L.R., Nagai S., Shimada K., Meister P., Taddei A., Gasser S.M.. Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr. Biol. 2011; 21:25–33. - PubMed
-
- Sinclair D.A., Guarente L.. Extrachromosomal rDNA circles - a cause of aging in yeast. Cell. 1997; 91:1033–1042. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

