Machine learning and artificial intelligence in physiologically based pharmacokinetic modeling
- PMID: 36156156
- PMCID: PMC9887681
- DOI: 10.1093/toxsci/kfac101
Machine learning and artificial intelligence in physiologically based pharmacokinetic modeling
Abstract
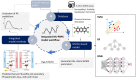
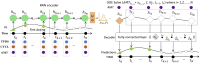
Physiologically based pharmacokinetic (PBPK) models are useful tools in drug development and risk assessment of environmental chemicals. PBPK model development requires the collection of species-specific physiological, and chemical-specific absorption, distribution, metabolism, and excretion (ADME) parameters, which can be a time-consuming and expensive process. This raises a need to create computational models capable of predicting input parameter values for PBPK models, especially for new compounds. In this review, we summarize an emerging paradigm for integrating PBPK modeling with machine learning (ML) or artificial intelligence (AI)-based computational methods. This paradigm includes 3 steps (1) obtain time-concentration PK data and/or ADME parameters from publicly available databases, (2) develop ML/AI-based approaches to predict ADME parameters, and (3) incorporate the ML/AI models into PBPK models to predict PK summary statistics (eg, area under the curve and maximum plasma concentration). We also discuss a neural network architecture "neural ordinary differential equation (Neural-ODE)" that is capable of providing better predictive capabilities than other ML methods when used to directly predict time-series PK profiles. In order to support applications of ML/AI methods for PBPK model development, several challenges should be addressed (1) as more data become available, it is important to expand the training set by including the structural diversity of compounds to improve the prediction accuracy of ML/AI models; (2) due to the black box nature of many ML models, lack of sufficient interpretability is a limitation; (3) Neural-ODE has great potential to be used to generate time-series PK profiles for new compounds with limited ADME information, but its application remains to be explored. Despite existing challenges, ML/AI approaches will continue to facilitate the efficient development of robust PBPK models for a large number of chemicals.
Keywords: in vitro to in vivo extrapolation (IVIVE); artificial intelligence; machine learning; pharmacometrics; physiologically based pharmacokinetic (PBPK) modeling; risk assessment.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society of Toxicology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Agatonovic-Kustrin S., Beresford R., Yusof A. P. M. (2001). Theoretically-derived molecular descriptors important in human intestinal absorption. J. Pharm. Biomed. Anal. 25, 227–237. - PubMed
-
- Andersen M. E., Mallick P., Clewell H. J. 3rd, Yoon M., Olsen G. W., Longnecker M. P. (2021). Using quantitative modeling tools to assess pharmacokinetic bias in epidemiological studies showing associations between biomarkers and health outcomes at low exposures. Environ. Res. 197, 111183. - PubMed
-
- Athersuch T. J., Wilson I. D., Keun H. C., Lindon J. C. (2013). Development of quantitative structure-metabolism (QSMR) relationships for substituted anilines based on computational chemistry. Xenobiotica 43, 792–802. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

