Feasibility of Automated Segmentation of Pigmented Choroidal Lesions in OCT Data With Deep Learning
- PMID: 36156729
- PMCID: PMC9526362
- DOI: 10.1167/tvst.11.9.25
Feasibility of Automated Segmentation of Pigmented Choroidal Lesions in OCT Data With Deep Learning
Abstract
Purpose: To evaluate the feasibility of automated segmentation of pigmented choroidal lesions (PCLs) in optical coherence tomography (OCT) data and compare the performance of different deep neural networks.
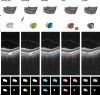
Methods: Swept-source OCT image volumes were annotated pixel-wise for PCLs and background. Three deep neural network architectures were applied to the data: the multi-dimensional gated recurrent units (MD-GRU), the V-Net, and the nnU-Net. The nnU-Net was used to compare the performance of two-dimensional (2D) versus three-dimensional (3D) predictions.
Results: A total of 121 OCT volumes were analyzed (100 normal and 21 PCLs). Automated PCL segmentations were successful with all neural networks. The 3D nnU-Net predictions showed the highest recall with a mean of 0.77 ± 0.22 (MD-GRU, 0.60 ± 0.31; V-Net, 0.61 ± 0.25). The 3D nnU-Net predicted PCLs with a Dice coefficient of 0.78 ± 0.13, outperforming MD-GRU (0.62 ± 0.23) and V-Net (0.59 ± 0.24). The smallest distance to the manual annotation was found using 3D nnU-Net with a mean maximum Hausdorff distance of 315 ± 172 µm (MD-GRU, 1542 ± 1169 µm; V-Net, 2408 ± 1060 µm). The 3D nnU-Net showed a superior performance compared with stacked 2D predictions.
Conclusions: The feasibility of automated deep learning segmentation of PCLs was demonstrated in OCT data. The neural network architecture had a relevant impact on PCL predictions.
Translational relevance: This work serves as proof of concept for segmentations of choroidal pathologies in volumetric OCT data; improvements are conceivable to meet clinical demands for the diagnosis, monitoring, and treatment evaluation of PCLs.
Conflict of interest statement
Disclosure:
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

