Full-length transcriptome revealed the accumulation of polyunsaturated fatty acids in developing seeds of Plukenetia volubilis
- PMID: 36157055
- PMCID: PMC9504451
- DOI: 10.7717/peerj.13998
Full-length transcriptome revealed the accumulation of polyunsaturated fatty acids in developing seeds of Plukenetia volubilis
Abstract
Background: Plukenetia volubilis is cultivated as a valuable oilseed crop, and its mature seeds are rich in polyunsaturated fatty acids (FAs), which are widely used in food and pharmaceutical industries. Recently, next-generation sequencing (NGS) transcriptome studies in P. volubilis indicated that some candidate genes were involved in oil biosynthesis. The NGS were inaccuracies in assembly of some candidate genes, leading to unknown errors in date analyses. However, single molecular real-time (SMRT) sequencing can overcome these assembled errors. Unfortunately, this technique has not been reported in P. volubilis.
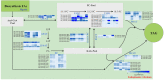
Methods: The total oil content of P. volubilis seed (PVS) was determined using Soxhlet extraction system. The FA composition were analyzed by gas chromatography. Combining PacBio SMRT and Illumina technologies, the transcriptome analysis of developing PVS was performed. Functional annotation and differential expression were performed by BLAST software (version 2.2.26) and RSEM software (version 1.2.31), respectively. The lncRNA-targeted transcripts were predicted in developing PVS using LncTar tool.
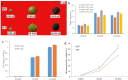
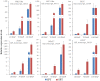
Results: By Soxhlet extraction system, the oil content of superior plant-type (SPT) was 13.47% higher than that of inferior plant-type (IPT) at mature PVS. The most abundant FAs were C18:2 and C18:3, among which C18:3 content of SPT was 1.11-fold higher than that of IPT. Combined with PacBio and Illumina platform, 68,971 non-redundant genes were obtained, among which 7,823 long non-coding RNAs (lncRNAs) and 7,798 lncRNA-targeted genes were predicted. In developing seed, the expressions of 57 TFs showed a significantly positive correlation with oil contents, including WRI1-like1, LEC1-like1, and MYB44-like. Comparative analysis of expression profiles between SPT and IPT implied that orthologs of FAD3, PDCT, PDAT, and DAGT2 were possibly important for the accumulation of polyunsaturated FAs. Together, these results provide a reference for oil biosynthesis of P. volubilis and genetic improvement of oil plants.
Keywords: Full-length transcriptome; Oil accumulation; Plukenetia volubilis; Polyunsaturated fatty acids; Transcription factors.
©2022 Fu et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



Similar articles
-
De novo transcriptome assembly of the eight major organs of Sacha Inchi (Plukenetia volubilis) and the identification of genes involved in α-linolenic acid metabolism.BMC Genomics. 2018 May 22;19(1):380. doi: 10.1186/s12864-018-4774-y. BMC Genomics. 2018. PMID: 29788925 Free PMC article.
-
Transcriptome analysis of Sacha Inchi (Plukenetia volubilis L.) seeds at two developmental stages.BMC Genomics. 2012 Dec 20;13:716. doi: 10.1186/1471-2164-13-716. BMC Genomics. 2012. PMID: 23256450 Free PMC article.
-
De novo transcriptome assembly and comparative analysis between male and benzyladenine-induced female inflorescence buds of Plukenetia volubilis.J Plant Physiol. 2018 Feb;221:107-118. doi: 10.1016/j.jplph.2017.12.006. Epub 2017 Dec 9. J Plant Physiol. 2018. PMID: 29275214
-
Sacha inchi (Plukenetia volubilis L.)-from lost crop of the Incas to part of the solution to global challenges?Planta. 2020 Mar 17;251(4):80. doi: 10.1007/s00425-020-03377-3. Planta. 2020. PMID: 32185506 Review.
-
Sacha inchi (Plukenetia volubilis L.): Nutritional composition, biological activity, and uses.Food Chem. 2018 Nov 1;265:316-328. doi: 10.1016/j.foodchem.2018.05.055. Epub 2018 May 29. Food Chem. 2018. PMID: 29884388 Review.
Cited by
-
Identification of the circRNA-miRNA-mRNA Regulatory Network in Pterygium-Associated Conjunctival Epithelium.Biomed Res Int. 2022 Nov 8;2022:2673890. doi: 10.1155/2022/2673890. eCollection 2022. Biomed Res Int. 2022. PMID: 36398070 Free PMC article.
-
N6-methyladenosine RNA methyltransferase CpMTA1 mediates CpAphA mRNA stability through a YTHDF1-dependent m6A modification in the chestnut blight fungus.PLoS Pathog. 2024 Aug 19;20(8):e1012476. doi: 10.1371/journal.ppat.1012476. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39159278 Free PMC article.
-
Autophagy mediated by ROS-AKT-FoxO pathway is required for intestinal regeneration in echinoderms.Cell Commun Signal. 2025 Jan 7;23(1):8. doi: 10.1186/s12964-024-01993-0. Cell Commun Signal. 2025. PMID: 39762855 Free PMC article.
-
The Evolution of Lipidomics during Oil Accumulation of Plukenetia volubilis Seeds.Plants (Basel). 2024 Aug 8;13(16):2193. doi: 10.3390/plants13162193. Plants (Basel). 2024. PMID: 39204629 Free PMC article.
References
-
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue GF, Drira N, Ohlrogge JB, Arondel V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12527–12532. doi: 10.1073/pnas.1106502108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

