Identification of uveitis-associated functions based on the feature selection analysis of gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment scores
- PMID: 36157069
- PMCID: PMC9493498
- DOI: 10.3389/fnmol.2022.1007352
Identification of uveitis-associated functions based on the feature selection analysis of gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment scores
Abstract
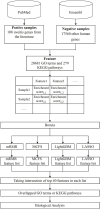
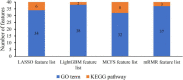
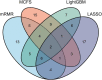
Uveitis is a typical type of eye inflammation affecting the middle layer of eye (i.e., uvea layer) and can lead to blindness in middle-aged and young people. Therefore, a comprehensive study determining the disease susceptibility and the underlying mechanisms for uveitis initiation and progression is urgently needed for the development of effective treatments. In the present study, 108 uveitis-related genes are collected on the basis of literature mining, and 17,560 other human genes are collected from the Ensembl database, which are treated as non-uveitis genes. Uveitis- and non-uveitis-related genes are then encoded by gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment scores based on the genes and their neighbors in STRING, resulting in 20,681 GO term features and 297 KEGG pathway features. Subsequently, we identify functions and biological processes that can distinguish uveitis-related genes from other human genes by using an integrated feature selection method, which incorporate feature filtering method (Boruta) and four feature importance assessment methods (i.e., LASSO, LightGBM, MCFS, and mRMR). Some essential GO terms and KEGG pathways related to uveitis, such as GO:0001841 (neural tube formation), has04612 (antigen processing and presentation in human beings), and GO:0043379 (memory T cell differentiation), are identified. The plausibility of the association of mined functional features with uveitis is verified on the basis of the literature. Overall, several advanced machine learning methods are used in the current study to uncover specific functions of uveitis and provide a theoretical foundation for the clinical treatment of uveitis.
Keywords: KEGG pathway; enrichment theory; feature selection; gene ontology; uveitis.
Copyright © 2022 Lu, Wang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Identification of protein-protein interaction associated functions based on gene ontology and KEGG pathway.Front Genet. 2022 Sep 12;13:1011659. doi: 10.3389/fgene.2022.1011659. eCollection 2022. Front Genet. 2022. PMID: 36171880 Free PMC article.
-
Analysis of Four Types of Leukemia Using Gene Ontology Term and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Scores.Comb Chem High Throughput Screen. 2020;23(4):295-303. doi: 10.2174/1386207322666181231151900. Comb Chem High Throughput Screen. 2020. PMID: 30599106
-
Analysis and prediction of protein stability based on interaction network, gene ontology, and KEGG pathway enrichment scores.Biochim Biophys Acta Proteins Proteom. 2023 May 1;1871(3):140889. doi: 10.1016/j.bbapap.2023.140889. Epub 2023 Jan 4. Biochim Biophys Acta Proteins Proteom. 2023. PMID: 36610583
-
Identification of Common Genes and Pathways in Eight Fibrosis Diseases.Front Genet. 2021 Jan 15;11:627396. doi: 10.3389/fgene.2020.627396. eCollection 2020. Front Genet. 2021. PMID: 33519923 Free PMC article.
-
The use of Gene Ontology terms and KEGG pathways for analysis and prediction of oncogenes.Biochim Biophys Acta. 2016 Nov;1860(11 Pt B):2725-34. doi: 10.1016/j.bbagen.2016.01.012. Epub 2016 Jan 20. Biochim Biophys Acta. 2016. PMID: 26801878
Cited by
-
Comparative Efficacy of Interleukin-7 and -15 Blockade in Alleviating Experimental Chronic Uveitis and Suppressing Pathogenic Memory CD4+ T Cells.Invest Ophthalmol Vis Sci. 2025 May 1;66(5):9. doi: 10.1167/iovs.66.5.9. Invest Ophthalmol Vis Sci. 2025. PMID: 40323267 Free PMC article.
-
Delineating the Acquired Genetic Diversity and Multidrug Resistance in Alcaligenes from Poultry Farms and Nearby Soil.J Microbiol. 2024 Jul;62(7):511-523. doi: 10.1007/s12275-024-00129-w. Epub 2024 Jun 21. J Microbiol. 2024. PMID: 38904697
References
-
- Chee S., Jap A. (2010). Cytomegalovirus anterior uveitis: outcome of treatment. Br. J. Ophthalmol. 94 1648–1652. - PubMed
LinkOut - more resources
Full Text Sources

