Predicting Prognosis of Hepatocellular Carcinoma Patients Based on the Expression Signatures of Mitophagy Genes
- PMID: 36157211
- PMCID: PMC9507775
- DOI: 10.1155/2022/4835826
Predicting Prognosis of Hepatocellular Carcinoma Patients Based on the Expression Signatures of Mitophagy Genes
Abstract
Background: The unbalance of mitophagy was closely related to hepatocellular carcinoma (HCC) progression. At present, it has not been uncovered about the influence of mitophagy genes on HCC prognosis and their potential pathogenesis.
Materials and methods: The expression and clinical information of HCC in TCGA cohort were used to identify mitophagy differentially expressed genes (MDEGs) with prognostic value. The prognostic model of mitophagy genes was built and externally validated by LASSO regression in TCGA cohort and ICGC cohort, respectively. The function of the prognostic signature and its association with immune cell infiltration were explored. The profile of MDEGs was validated with 39 pairs HCC and paracarcinoma tissues by quantitative reverse transcription-PCR (qRT-PCR).
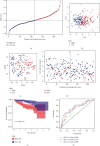
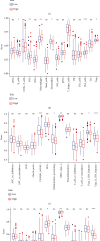
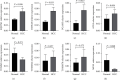
Results: A total of 18 mitophagy genes that were upregulated and contributed to poor prognosis in HCC were identified. These genes could interact with each other. The correlation analysis showed that there was positively correlation among mitophagy genes. According to optimal λ value, 8 mitophagy gene signatures were involved in prognostic model. Based on median risk scores, HCC patients were divided into high-risk group and low-risk group. Compared with the low-risk group, the high-risk group has worse overall survival in TCGA cohort and ICGC cohort. The univariate and multivariate Cox regression analysis suggested that risk score was an independent prognostic factor of HCC patients. Time-dependent ROC curve was used to identify and validate good predicting performance of the prognostic model. Enrichment analysis showed that risk differentially expressed genes were enriched in various metabolism and cell division processes. The immune cell infiltration score and immune function were significantly different in two groups. qRT-PCR validation result showed that QSTM1, CSNK2B, PGAM5, and ATG5 were upregulated.
Conclusion: Mitophagy genes could influence HCC progression through regulating the metabolism and immune functions and could be used to predict prognosis and considered as potential prognostic biomarker and precise therapeutic target of HCC.
Copyright © 2022 Yan-ke Li et al.
Conflict of interest statement
All authors disclose no conflicts of interest that might bias their work.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

