Gibberellin-regulated proteins: Emergent allergens
- PMID: 36157274
- PMCID: PMC9500206
- DOI: 10.3389/falgy.2022.877553
Gibberellin-regulated proteins: Emergent allergens
Abstract
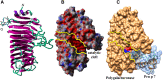
About 10 years ago, a protein family was shown for the first time to contain allergenic members, gibberellin-regulated protein (GRP). The first reported member was from peach, Pru p 7. One can hypothesize that it was not detected before because its physicochemical characteristics overlap with those of lipid transfer protein (LTP), a well-known allergen, or because the exposure to GRP increased due to an increase in the gibberellin phythormone level in plant food, either exogenous or endogenous. Like LTPs, GRPs are small cationic proteins with disulfide bridges, are resistant to heat and proteolytic cleavage, and are involved in the defense of the plant. Besides peach, GRP allergens have been described in Japanese apricot (Pru m 7), sweet cherry (Pru av 7), orange (Cit s 7), pomegranate (Pun g 7), bell pepper (Cap a 7), strawberry (Fra a GRP), and also in pollen with a restriction to Cupressaceae tree family (Cup s 7, Cry j 7, and Jun a 7). IgE cross-reactivities were described between GRPs, and the reported peach/cypress and citrus/cypress syndromes may therefore be explained because of these GRP cross-reactivities. GRPs are clinically relevant, and severe adverse reactions may sometimes occur in association with cofactors. More than 60% and up to 95% sequence identities are calculated between various allergenic GRPs, and three-dimensional models show a cleft in the molecule and predict at least three epitopic regions. The structure of the protein and its properties and the matrix effect in the original allergenic source should be unraveled to understand why, despite the ubiquity of the protein family in plants, only a few members are able to sensitize patients.
Keywords: 3D structure; food allergy; gibberellin-regulated protein; pollen allergy; pollen food allergy syndrome.
© 2022 Iizuka, Barre, Rougé, Charpin, Scala, Baudin, Aizawa, Sénéchal and Poncet.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

