Local inflammatory response to gastroesophageal reflux: Association of gene expression of inflammatory cytokines with esophageal multichannel intraluminal impedance-pH data
- PMID: 36159432
- PMCID: PMC9477692
- DOI: 10.12998/wjcc.v10.i26.9254
Local inflammatory response to gastroesophageal reflux: Association of gene expression of inflammatory cytokines with esophageal multichannel intraluminal impedance-pH data
Abstract
Background: Gene expression of inflammatory cytokines may take part in the pathophysiology of different forms of gastroesophageal reflux disease (GERD).
Aim: To explore gene expression of inflammatory cytokines in esophageal mucosa in patients with erosive esophagitis (EE) and non-erosive forms of GERD (NERD) and its association with data of esophageal multichannel intraluminal impedance-pH (MII-pH) measurements.
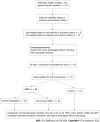
Methods: This was a single-center prospective study. Esophageal mucosa samples were taken from the lower part of the esophagus during endoscopy. Expression of interleukin (IL)-1β, IL-10, IL-18, tumor necrosis factor α (TNFA), toll-like receptor 4 (TLR4), GATA binding protein 3 (GATA3), differentiation cluster 68 (CD68) and β-2 microglobulin genes in esophageal mucosa was assessed with ImmunoQuantex assays. MII-pH measurements were performed on all the participants. Diagnosis of GERD was confirmed by the results of the MII-pH data. Based on the endoscopy, patients were allocated to the groups of EE and NERD. The control group consisted of non-symptomatic subjects with normal endoscopy and MII-pH results. We used nonparametric statistics to compare the differences between the groups. Association of expression of the mentioned genes with the results of the MII-pH data was assessed with Spearman's rank method.
Results: Data from 60 patients with GERD and 10 subjects of the control group were available for the analysis. Higher expression of IL-18 (5.89 ± 0.4 vs 5.28 ± 1.1, P = 0.04) and GATA3 (2.92 ± 0.86 vs 2.23 ± 0.96, P = 0.03) was found in the EE group compared to NERD. Expression of IL-1β, IL-18, TNFA, and TLR4 was lower (P < 0.05) in the control group compared to EE and NERD. Esophageal acid exposure correlated with the expression of IL-1β (Spearman's rank r = 0.29), IL-18 (r = 0.31), TNFA (r = 0.35), GATA3 (r = 0.34), TLR4 (r = 0.29), and CD68 (r = 0.37). Mean esophageal рН correlated inversely with the expression of IL-18, TNFA, GATA3, TLR4, and CD68. No association of gene expression with the number of gastroesophageal refluxes was found.
Conclusion: In patients with EE, local expression of IL-18 and GATA3 was higher compared to subjects with NERD. Esophageal acid exposure correlated directly with expression of IL-1β, IL-18, TNFA, TLR4, CD68, and β-2 microglobulin genes. Inverse correlation was revealed between expression of IL-18, TNFA, GATA3, TLR4, and CD68 and mean esophageal pH.
Keywords: Cytokines; Erosive esophagitis; Esophageal multichannel intraluminal impedance-pH; Gastroesophageal reflux; Gastroesophageal reflux disease; Gene expression; Non-erosive gastroesophageal reflux disease.
©The Author(s) 2022. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: Dr. Morozov reports grants from Russian Science Foundation, personal fees from AstraZeneca, personal fees from AlfaSigma, non-financial support from Laborie, personal fees from DrFalk, personal fees from Takeda, from Federal Research Center of Nutrition and Biotechnology, outside the submitted work.
Figures
Similar articles
-
How many cases of laryngopharyngeal reflux suspected by laryngoscopy are gastroesophageal reflux disease-related?World J Gastroenterol. 2012 Aug 28;18(32):4363-70. doi: 10.3748/wjg.v18.i32.4363. World J Gastroenterol. 2012. PMID: 22969200 Free PMC article.
-
Acid and non-acid reflux patterns in patients with erosive esophagitis and non-erosive reflux disease (NERD): a study using intraluminal impedance monitoring.Dig Dis Sci. 2008 Jun;53(6):1506-12. doi: 10.1007/s10620-007-0059-z. Dig Dis Sci. 2008. PMID: 17934853
-
Reflux profile of Chinese gastroesophageal reflux disease patients with combined multichannel intraluminal impedance-pH monitoring.J Gastroenterol Hepatol. 2009 Jun;24(6):1113-8. doi: 10.1111/j.1440-1746.2009.05861.x. J Gastroenterol Hepatol. 2009. PMID: 19638089
-
Indications of 24-h esophageal pH monitoring, capsule pH monitoring, combined pH monitoring with multichannel impedance, esophageal manometry, radiology and scintigraphy in gastroesophageal reflux disease?Turk J Gastroenterol. 2017 Dec;28(Suppl 1):S16-S21. doi: 10.5152/tjg.2017.06. Turk J Gastroenterol. 2017. PMID: 29199161 Review.
-
Evaluation of Esophageal Mucosal Integrity in Patients with Gastroesophageal Reflux Disease.Digestion. 2018;97(1):31-37. doi: 10.1159/000484106. Epub 2018 Feb 1. Digestion. 2018. PMID: 29393167 Review.
Cited by
-
Causal relationships between 91 inflammatory cytokines and gastroesophageal reflux disease, and the mediating role of related metabolites: Evidence from genetics.Medicine (Baltimore). 2025 May 30;104(22):e42426. doi: 10.1097/MD.0000000000042426. Medicine (Baltimore). 2025. PMID: 40441226 Free PMC article.
-
Causal relationship between air pollution, lung function, gastroesophageal reflux disease, and non-alcoholic fatty liver disease: univariate and multivariate Mendelian randomization study.Front Public Health. 2024 Apr 29;12:1368483. doi: 10.3389/fpubh.2024.1368483. eCollection 2024. Front Public Health. 2024. PMID: 38746002 Free PMC article.
-
Esophageal Mucosal Resistance in Reflux Esophagitis: What We Have Learned So Far and What Remains to Be Learned.Diagnostics (Basel). 2023 Aug 12;13(16):2664. doi: 10.3390/diagnostics13162664. Diagnostics (Basel). 2023. PMID: 37627923 Free PMC article. Review.
-
Multidimensional mechanisms and therapies underlying gastroesophageal reflux disease: focus on immunity, signaling pathways, and the microbiota-gut-brain axis.Front Immunol. 2025 Jul 18;16:1629944. doi: 10.3389/fimmu.2025.1629944. eCollection 2025. Front Immunol. 2025. PMID: 40755761 Free PMC article. Review.
-
Causal association between gastroesophageal reflux disease and sepsis, and the mediating role of gut bacterial abundance, a Mendelian randomization study.Medicine (Baltimore). 2025 Feb 21;104(8):e41631. doi: 10.1097/MD.0000000000041631. Medicine (Baltimore). 2025. PMID: 39993106 Free PMC article.
References
-
- Vakil N, van Zanten SV, Kahrilas P, Dent J, Jones R Global Consensus Group. The Montreal definition and classification of gastroesophageal reflux disease: a global evidence-based consensus. Am J Gastroenterol. 2006;101:1900–20; quiz 1943. - PubMed
-
- Iwakiri K, Fujiwara Y, Manabe N, Ihara E, Kuribayashi S, Akiyama J, Kondo T, Yamashita H, Ishimura N, Kitasako Y, Iijima K, Koike T, Omura N, Nomura T, Kawamura O, Ohara S, Ozawa S, Kinoshita Y, Mochida S, Enomoto N, Shimosegawa T, Koike K. Evidence-based clinical practice guidelines for gastroesophageal reflux disease 2021. J Gastroenterol. 2022;57:267–285. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

