Gut and oral microbiota associations with viral mitigation behaviors during the COVID-19 pandemic
- PMID: 36159641
- PMCID: PMC9500509
- DOI: 10.3389/fcimb.2022.966361
Gut and oral microbiota associations with viral mitigation behaviors during the COVID-19 pandemic
Abstract
Imposition of social and health behavior mitigations are important control measures in response to the coronavirus disease 2019 (COVID-19) pandemic caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Although postulated that these measures may impact the human microbiota including losses in diversity from heightened hygiene and social distancing measures, this hypothesis remains to be tested. Other impacts on the microbiota and host mental and physical health status associations from these measures are also not well-studied. Here we examine changes in stool and oral microbiota by analyzing 16S rRNA gene sequence taxonomic profiles from the same individuals during pre-pandemic (before March 2020) and early pandemic (May-November 2020) phases. During the early pandemic phase, individuals were also surveyed using questionnaires to report health histories, anxiety, depression, sleep and other lifestyle behaviors in a cohort of predominantly Caucasian adults (mean age = 61.5 years) with the majority reporting at least one underlying co-morbidity. We identified changes in microbiota (stool n = 288; oral n = 89) between pre-pandemic and early pandemic time points from the same subject and associated these differences with questionnaire responses using linear statistical models and hierarchical clustering of microbiota composition coupled to logistic regression. While a trend in loss of diversity was identified between pre-pandemic and early pandemic time points it was not statistically significant. Paired difference analyses between individuals identified fewer significant changes between pre-pandemic and early pandemic microbiota in those who reported fewer comorbidities. Cluster transition analyses of stool and saliva microbiota determined most individuals remained in the same cluster assignments from the pre-pandemic to early pandemic period. Individuals with microbiota that shifted in composition, causing them to depart a pre-pandemic cluster, reported more health issues and pandemic-associated worries. Collectively, our study identified that stool and saliva microbiota from the pre-pandemic to early pandemic periods largely exhibited ecological stability (especially stool microbiota) with most associations in loss of diversity or changes in composition related to more reported health issues and pandemic-associated worries. Longitudinal observational cohorts are necessary to monitor the microbiome in response to pandemics and changes in public health measures.
Keywords: 16S rRNA gene amplicon sequencing; COVID-19; ecological stability; gut microbiota; microbiome; saliva microbiota.
Copyright © 2022 Li, Methé, Fitch, Gentry, Kessinger, Patel, Petraglia, Swamy and Morris.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
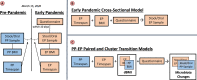
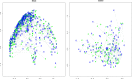

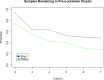
References
-
- Aitchison J. (1982). The statistical analysis of compositional data. J. R. Stat. Society: Ser. B (Methodological) 44 (2), 139–160. doi: 10.1111/j.2517-6161.1982.tb01195.x - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

