A specific immune signature for predicting the prognosis of glioma patients with IDH1-mutation and guiding immune checkpoint blockade therapy
- PMID: 36159801
- PMCID: PMC9500319
- DOI: 10.3389/fimmu.2022.1001381
A specific immune signature for predicting the prognosis of glioma patients with IDH1-mutation and guiding immune checkpoint blockade therapy
Abstract
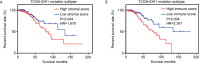
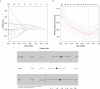
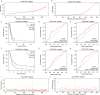
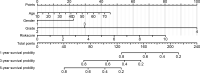
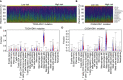
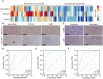
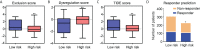
Isocitrate dehydrogenase (IDH1) is frequently mutated in glioma tissues, and this mutation mediates specific tumor-promoting mechanisms in glioma cells. We aimed to identify specific immune biomarkers for IDH1-mutation (IDH1mt) glioma. The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) were used to obtain RNA sequencing data and clinical characteristics of glioma tissues, while the stromal and immune scores of TCGA glioma tissues were determined using the ESTIMATE algorithm. Differentially expressed genes (DEGs), the protein-protein interaction(PPI) network, and least absolute shrinkage and selection operator (LASSO) and Cox regression analyses were used to select hub genes associated with stroma and immune scores and the prognoses of patients and to construct the risk model. The practicability and specificity of the risk model in both IDH1mt and IDH1-wildtype (wtIDH1) gliomas in TCGA and CGGA were evaluated. Molecular mechanisms, immunological characteristics and benefits of immune checkpoint blockade therapy in glioma tissues with IDH1mt were analyzed using GSEA, immunohistochemical staining, CIBERSORT, and T-cell dysfunction and exclusion (TIDE) analysis. The overall survival rate for IDH1mt-glioma patients with high stroma/immune scores was lower than that for those with low stroma/immune scores. A total of 222 DEGs were identified in IDH1mt glioma tissues with high stroma/immune scores. Among them, 72 genes had interactions in the PPI network, while three genes, HLA-DQA2, HOXA3, and SAA2, were selected as hub genes and used to construct risk models classifying patients into high- and low-risk score groups, followed by LASSO and Cox regression analyses. This risk model showed prognostic value in IDH1mt glioma in both TCGA and CCGA; nevertheless, the model was not suitable for wtIDH1 glioma. The risk model may act as an independent prognostic factor for IDH1mt glioma. IDH1mt glioma tissues from patients with high-risk scores showed more infiltration of M1 and CD8 T cells than those from patients with low-risk scores. Moreover, TIDE analysis showed that immune checkpoint blockade(ICB) therapy was highly beneficial for IDH1mt patients with high-risk scores. The risk model showed specific potential to predict the prognosis of IDH1mt-glioma patients, as well as guide ICB, contributing to the diagnosis and therapy of IDH1mt-glioma patients.
Keywords: IDH1 mutation; glioma; immune; immune checkpoint blockade therapy; signature.
Copyright © 2022 Zeng, Hu, Ruan, Zhang, Lei, Yang, Peng, Pan and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

