Genomic selection in United States dairy cattle
- PMID: 36159997
- PMCID: PMC9500184
- DOI: 10.3389/fgene.2022.994466
Genomic selection in United States dairy cattle
Abstract
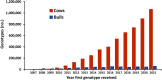
The genomic selection program for dairy cattle in the United States has doubled the rate of genetic gain. Since 2010, the average annual increase in net merit has been $85 compared to $40 during the previous 5 years. The number of genotypes has been rapidly increasing both domestically and internationally and reached over 6.5 million in 2022 with 1,134,593 submitted in 2021. Evaluations are calculated for over 50 traits. Feed efficiency (residual feed intake), heifer and cow livability, age at first calving, six health traits, and gestation length have been added in recent years to represent the economic value of selection candidates more accurately; work is underway to develop evaluations for hoof health. Evaluations of animals with newly submitted genotypes are calculated weekly. In April 2019, evaluations were extended to crossbreds; to support that effort, evaluations are initially calculated on an all-breed base and then blended by an estimated breed composition. For animals that are less than 90% of one breed, the evaluation is calculated by weighting contributions of each of the five major dairy breeds evaluated (Ayrshire, Brown Swiss, Guernsey, Holstein, and Jersey) by the breed proportion. Nearly 200,000 animals received blended evaluations in July 2022. Pedigree is augmented by using haplotype matching to discover maternal grandsires and great-grandsires. Haplotype analysis is also used to discover undesirable recessive conditions. In many cases, the causative variant has been identified, and results from a gene test or inclusion on a genotyping chip improves the accuracy of those determinations for the current 27 conditions reported. Recently discovered recessive conditions include neuropathy with splayed forelimbs in Jerseys, early embryonic death in Holsteins, and curly calves in Ayrshires. Techniques have been developed to support rapid searches for parent-progeny relationships and identical genotypes among all likely genotypes, which substantially reduces processing time. Work continues on using sequence data to discover additional informative single nucleotide polymorphisms and to incorporate those previously discovered. Adoption of genotyping by sequencing is expected to improve flexibility of marker selection. The success of the Council on Dairy Cattle Breeding in conducting the genetic evaluation program is the result of close cooperation with industry and research groups, including the United States Department of Agriculture, breed associations, genotyping laboratories, and artificial-insemination organizations.
Keywords: ancestor discovery; breed composition; dairy cattle; genetic gain; genetic-economic index; genomic selection; haplotype; recessive discovery.
Copyright © 2022 Wiggans and Carrillo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Cole J. B., VanRaden P. M., Null D. J., Hutchison J. L., Hubbard S. M. (2022). Haplotype tests for economically important traits of dairy cattle. AIP Res. Rep. Genomic5 (12-20). Available at: https://www.ars.usda.gov/ARSUserFiles/80420530/Publications/ARR/Haplotyp.... Accessed date July 10, 2022.
-
- Council on Dairy Cattle Breeding (2020). Feed saved (FSAV). Trait reference sheet. Available at: https://www.uscdcb.com/wp-content/uploads/2020/11/CDCB-Reference-Sheet-F.... Accessed July 13, 2022.
-
- Council on Dairy Cattle Breeding (2022a). Genetic evaluations. Available at: https://www.uscdcb.com/what-we-do/genetic-evaluations/. Accessed date July 13, 2022.
-
- Council on Dairy Cattle Breeding (2022b). Genotype counts by chip type, breed code, and sex code in database as of 2022-06-27. Available at: https://queries.uscdcb.com/Genotype/cur_freq.html. Accessed date June 30, 2022.
-
- Holstein Association USA (2022). Interpreting linear type trait STAs. Available at: https://www.holsteinusa.com/genetic_evaluations/ss_interpret_linear.html. Accessed date July 13, 2022.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

