Ascertaining yield and grain protein content stability in wheat genotypes having the Gpc-B1 gene using univariate, multivariate, and correlation analysis
- PMID: 36160017
- PMCID: PMC9490372
- DOI: 10.3389/fgene.2022.1001904
Ascertaining yield and grain protein content stability in wheat genotypes having the Gpc-B1 gene using univariate, multivariate, and correlation analysis
Abstract
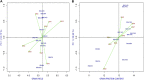
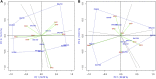
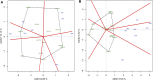
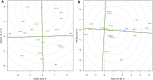
The high performance and stability of wheat genotypes for yield, grain protein content (GPC), and other desirable traits are critical for varietal development and food and nutritional security. Likewise, the genotype by environment (G × E) interaction (GEI) should be thoroughly investigated and favorably utilized whenever genotype selection decisions are made. The present study was planned with the following two major objectives: 1) determination of GEI for some advanced wheat genotypes across four locations (Ludhiana, Ballowal, Patiala, and Bathinda) of Punjab, India; and 2) selection of the best genotypes with high GPC and yield in various environments. Different univariate [Eberhart and Ruessll's models; Perkins and Jinks' models; Wrike's Ecovalence; and Francis and Kannenberg's models], multivariate (AMMI and GGE biplot), and correlation analyses were used to interpret the data from the multi-environmental trial (MET). Consequently, both the univariate and multivariate analyses provided almost similar results regarding the top-performing and stable genotypes. The analysis of variance revealed that variation due to environment, genotype, and GEI was highly significant at the 0.01 and 0.001 levels of significance for all studied traits. The days to flowering, plant height, spikelets per spike, grain per spike, days to maturity, and 1000-grain weight were specifically affected by the environment, whereas yield was mainly affected by the environment and GEI. Genotypes, on the other hand, had a greater impact on the GPC than environmental conditions. As a result, a multi-environmental investigation was necessary to identify the GEI for wheat genotype selection because the GEI was very significant for all of the evaluated traits. Yield, 1000-grain weight, spikelet per spike, and days to maturity were observed to have positive correlations, implying the feasibility of their simultaneous selection for yield enhancement. However, GPC was observed to have a negative correlation with yield. Patiala was found to be the most discriminating environment for both yield and GPC and also the most effective representative environment for GPC, whereas Ludhiana was found to be the most effective representative environment for yield. Eventually, two NILs (BWL7508, and BWL7511) were selected as the top across all environments for both yield and GPC.
Keywords: G × E interaction; grain protein content; multivariate analysis; stability analysis; univariate analysis; wheat.
Copyright © 2022 Tanin, Sharma, Saini, Singh, Kashyap, Srivastava, Mavi, Kaur, Kumar, Kumar, Grover, Chhuneja and Sohu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Ahakpaz F., Abdi H., Neyestani E., Hesami A., Mohammadi B., Mahmoudi K. N., et al. (2021). Genotype-by-environment interaction analysis for grain yield of barley genotypes under dryland conditions and the role of monthly rainfall. Agric. Water Manag. 245, 1–9. 10.1016/j.agwat.2020.106665 - DOI
-
- Ahmed Z. U., Panaullah G. M., Gauch H., McCouch S. R., Tyagi W., Kabir M. S., et al. (2011). Genotype and environment effects on rice (Oryza sativa L.) grain arsenic concentration in Bangladesh. Plant Soil 338, 367–382. 10.1007/s11104-010-0551-7 - DOI
-
- Amare K., Zeleke H., Bultosa G. (2015). Variability for yield, yield related traits and association among traits of sorghum (Sorghum Bicolor (L.) Moench) varieties in Wollo, Ethiopia. J. Plant Breed. Crop Sci. 7, 125–133. 10.5897/JPBCS2014.0469 - DOI
-
- Balakrishnan D., Subrahmanyam D., Badri J., Raju A. K., Rao Y. V., Beerelli K., et al. (2016). Genotype × environment interactions of yield traits in backcross introgression lines derived from Oryza sativa cv. Swarna/Oryza nivara . Front. Plant Sci. 7, 1–19. 10.3389/fpls.2016.01530 - DOI - PMC - PubMed
-
- Bantayehu M., Zelleke H., Assefa A. (2011). Genotype by environment interactions (G x E) and stability analyses of malting Barley (Hordeum distichon L.) genotypes across northwestern Ethiopia. Ethiop. J. Agric. Sci. 21 (1-2), 168–178.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

