Discovery of novel IDH1-R132C inhibitors through structure-based virtual screening
- PMID: 36160383
- PMCID: PMC9491111
- DOI: 10.3389/fphar.2022.982375
Discovery of novel IDH1-R132C inhibitors through structure-based virtual screening
Abstract
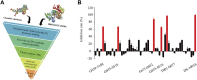
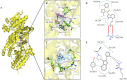
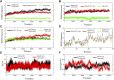
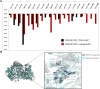
Isocitrate dehydrogenase (IDH) belongs to a family of enzymes involved in glycometabolism. It is found in many living organisms and is one of the most mutated metabolic enzymes. In the current study, we identified novel IDH1-R132C inhibitors using docking-based virtual screening and cellular inhibition assays. A total of 100 molecules with high docking scores were obtained from docking-based virtual screening. The cellular inhibition assay demonstrated five compounds at a concentration of 10 μM could inhibit cancer cells harboring the IDH1-R132C mutation proliferation by > 50%. The compound (T001-0657) showed the most potent effect against cancer cells harboring the IDH1-R132C mutation with a half-maximal inhibitory concentration (IC50) value of 1.311 μM. It also showed a cytotoxic effect against cancer cells with wild-type IDH1 and normal cells with IC50 values of 49.041 μM and >50 μM, respectively. Molecular dynamics simulations were performed to investigate the stability of the kinase structure binding of allosteric inhibitor compound A and the identified compound T001-0657 binds to IDH1-R132C. Root-mean-square deviation, root-mean-square fluctuation, and binding free energy calculations showed that both compounds bind tightly to IDH1-R132C. In conclusion, the compound identified in this study had high selectivity for cancer cells harboring IDH1-R132C mutation and could be considered a promising hit compound for further development of IDH1-R132C inhibitors.
Keywords: IDH1-R132C inhibitor; molecular docking; molecular dynamics simulation; tricarboxylic acid cycle (TCA cycle); virtual screening.
Copyright © 2022 Hu, Zeng, Ma, Yin, Zhao, Chen, Tang and Zuo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abbas S., Lugthart S., Kavelaars F. G., Schelen A., Koenders J. E., Zeilemaker A., et al. (2010). Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: Prevalence and prognostic value. Blood 116, 2122–2126. 10.1182/blood-2009-11-250878 PubMed Abstract | 10.1182/blood-2009-11-250878 | Google Scholar - DOI - DOI - PubMed
-
- Alhossary A., Handoko S. D., Mu Y., Kwoh C. K. (2015). Fast, accurate, and reliable molecular docking with QuickVina 2. Bioinformatics 31, 2214–2216. 10.1093/bioinformatics/btv082 PubMed Abstract | 10.1093/bioinformatics/btv082 | Google Scholar - DOI - DOI - PubMed
-
- Amary M. F., Bacsi K., Maggiani F., Damato S., Halai D., Berisha F., et al. (2011). IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 224, 334–343. 10.1002/path.2913 PubMed Abstract | 10.1002/path.2913 | Google Scholar - DOI - DOI - PubMed
-
- Amatangelo M. D., Quek L., Shih A., Stein E. M., Roshal M., David M. D., et al. (2017). Enasidenib induces acute myeloid leukemia cell differentiation to PromoteClinical response. Blood 130, 732–741. 10.1182/blood-2017-04-779447 PubMed Abstract | 10.1182/blood-2017-04-779447 | Google Scholar - DOI - DOI - PMC - PubMed
-
- Badur M. G., Muthusamy T., Parker S. J., Ma S., McBrayer S. K., Cordes T., et al. (2018). Oncogenic R132 IDH1 mutations limit NADPH for de novo lipogenesis through (D)2-hydroxyglutarate production in fibrosarcoma sells. Cell Rep. 25, 1018–1026. e4. 10.1016/j.celrep.2018.09.074 PubMed Abstract | 10.1016/j.celrep.2018.09.074 | Google Scholar - DOI - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

