mRNA and lncRNA expression profiles of liver tissues in children with biliary atresia
- PMID: 36160912
- PMCID: PMC9468840
- DOI: 10.3892/etm.2022.11571
mRNA and lncRNA expression profiles of liver tissues in children with biliary atresia
Abstract
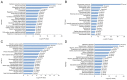
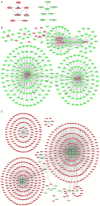
Progressive liver fibrosis is the most common phenotype in biliary atresia (BA). A number of pathways contribute to the fibrosis process so comprehensive understanding the mechanisms of liver fibrosis in BA will pave the way to improve patient's outcome after operation. In this study, the differentially expressed profiles of mRNAs and long non-coding RNAs from BA and choledochal cyst (CC) liver tissues were investigated and analyzed, which may provide potential clues to clarify hepatofibrosis mechanism in BA. A total of two BA and two CC liver tissue specimens were collected, the expression level of mRNAs and lncRNAs was detected by RNA sequencing. Differentially expressed mRNAs (DEmRNAs) were functionally annotated and protein-protein interaction networks (PPI) was established to predict the biological roles and interactive relationships. Differentially expressed lncRNAs (DElncRNAs) nearby targeted DEmRNA network and DElncRNA-DEmRNA co-expression network were constructed to further explore the roles of DElncRNAs in BA pathogenesis. The expression profiles of significant DEmRNAs were validated in Gene Expression Omnibus database. A total of 2,086 DEmRNAs and 184 DElncRNAs between BA and CC liver tissues were obtained. DEmRNAs were enriched in 521 Gene Ontology terms and 71 Kyoto Encyclopedia of Genes and Genomes terms which were mainly biological processes and metabolic pathways related to immune response and inflammatory response. A total of five hub proteins (TYRO protein tyrosine kinase binding protein, C-X-C motif chemokine ligand 8, pleckstrin, Toll-like receptor 8 and C-C motif chemokine receptor 5) were found in the PPI networks. A total of 31 DElncRNA-nearby-targeted DEmRNA pairs and 2,337 DElncRNA-DEmRNA co-expression pairs were obtained. The expression of DEmRNAs obtained from RNA sequencing were verified in GSE46960 dataset, generally. The present study identified key genes and lncRNAs participated in BA associated liver fibrosis, which may present a new avenue for understanding the patho-mechanism for hepatic fibrosis in BA.
Keywords: biliary atresia; bioinformation analysis; liver fibrosis; long non-coding RNA; mRNA.
Copyright: © Wu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Screening key lncRNAs for human rectal adenocarcinoma based on lncRNA-mRNA functional synergistic network.Cancer Med. 2019 Jul;8(8):3875-3891. doi: 10.1002/cam4.2236. Epub 2019 May 22. Cancer Med. 2019. PMID: 31116002 Free PMC article.
-
An integrative analysis of an lncRNA-mRNA competing endogenous RNA network to identify functional lncRNAs in uterine leiomyomas with RNA sequencing.Front Genet. 2023 Jan 4;13:1053845. doi: 10.3389/fgene.2022.1053845. eCollection 2022. Front Genet. 2023. PMID: 36685910 Free PMC article.
-
Identification of differentially expressed lncRNAs and mRNAs in luminal-B breast cancer by RNA-sequencing.BMC Cancer. 2019 Dec 3;19(1):1171. doi: 10.1186/s12885-019-6395-5. BMC Cancer. 2019. PMID: 31795964 Free PMC article.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
-
A narrative review of genes associated with liver fibrosis in biliary atresia.Transl Pediatr. 2024 Aug 31;13(8):1469-1478. doi: 10.21037/tp-24-94. Epub 2024 Aug 28. Transl Pediatr. 2024. PMID: 39263291 Free PMC article. Review.
Cited by
-
CircUTRN24/miR-483-3p/IGF-1 Regulates Autophagy Mediated Liver Fibrosis in Biliary Atresia.Mol Biotechnol. 2024 Jun;66(6):1424-1433. doi: 10.1007/s12033-023-00802-2. Epub 2023 Jun 27. Mol Biotechnol. 2024. PMID: 37369954
-
Liver-specific lncRNAs associated with liver cancers.FEBS Open Bio. 2025 Sep;15(9):1383-1405. doi: 10.1002/2211-5463.70079. Epub 2025 Jul 1. FEBS Open Bio. 2025. PMID: 40596757 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
