MsTHI1 overexpression improves drought tolerance in transgenic alfalfa (Medicago sativa L.)
- PMID: 36160983
- PMCID: PMC9495609
- DOI: 10.3389/fpls.2022.992024
MsTHI1 overexpression improves drought tolerance in transgenic alfalfa (Medicago sativa L.)
Abstract
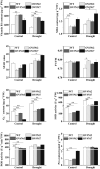
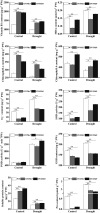
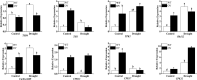
In recent years, drought stress caused by global warming has become a major constraint on agriculture. The thiamine thiazole synthase (THI1) is responsible for controlling thiamine production in plants displaying a response to various abiotic stresses. Nonetheless, most of the THI1 activities in plants remain largely unknown. In this study, we extracted MsTHI1 from alfalfa and demonstrated its beneficial impact on improving the resistance of plants to stress conditions. The highest levels of MsTHI1 expression were identified in alfalfa leaves, triggered by exposure to cold, drought, salt, or alkaline conditions. The upregulation of MsTHI1 in drought-stressed transgenic plants resulted in enhanced accumulation of vitamin B1 (VB1), chlorophyll a (Chl a), chlorophyll b (Chl b), soluble protein, higher soil and plant analyzer development (SPAD) value, and the activity of peroxidase (POD), maintained Fv/Fm, and decreased lipid peroxidation. Moreover, overexpression of MsTHI1 upregulated the transcription of THI4, TPK1, RbcX2, Cu/Zn-SOD, CPK13, and CPK32 and downregulated the transcription of TH1 and CPK17 in transgenic alfalfa under drought stress. These results suggested that MsTHI1 enhances drought tolerance by strengthening photosynthesis, regulating the antioxidant defense system, maintaining osmotic homeostasis, and mediating plant signal transduction.
Keywords: MsTHI1; alfalfa; antioxidant defense; drought tolerance; osmotic homeostasis; photosynthesis.
Copyright © 2022 Yin, Wang, Li, Zhang, Yang, Cui and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
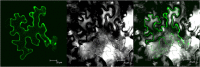


References
-
- Abidin A. A. Z., Yee W. S., Rahman N. S. A., Idris Z. H. C., Yusof Z. N. B. (2016). Osmotic, oxidative and salinity stresses upregulate the expressions of Thiamine (vitamin B1) biosynthesis genes (THIC and THI1/THI4) in oil palm (Elaeis guineensis). J. Oil Palm Res. 28, 308–319. 10.21894/jopr.2016.2803.07 - DOI
-
- Amjad S. F., Mansoora N., Yaseen S., Kamal A., Butt B., Matloob H., et al. (2021). Combined use of endophytic bacteria and pre-sowing treatment of thiamine mitigates the adverse effects of drought stress in wheat (Triticum aestivum L.) Cultivars. Sustainability 13, 6582. 10.3390/su13126582 - DOI
LinkOut - more resources
Full Text Sources

