Functional analysis of PHYB polymorphisms in Arabidopsis thaliana collected in Patagonia
- PMID: 36161012
- PMCID: PMC9490419
- DOI: 10.3389/fpls.2022.952214
Functional analysis of PHYB polymorphisms in Arabidopsis thaliana collected in Patagonia
Abstract
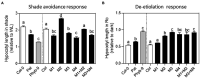
Arabidopsis thaliana shows a wide range of natural genetic variation in light responses. Shade avoidance syndrome is a strategy of major adaptive significance that includes seed germination, elongation of vegetative structures, leaf hyponasty, and acceleration of flowering. Previously, we found that the southernmost Arabidopsis accession, collected in the south of Patagonia (Pat), is hyposensitive to light and displays a reduced response to shade light. This work aimed to explore the genetic basis of the shade avoidance response (SAR) for hypocotyl growth by QTL mapping in a recently developed 162 RIL population between Col-0 and Pat. We mapped four QTL for seedling hypocotyl growth: WL1 and WL2 QTL in white light, SHADE1 QTL in shade light, and SAR1 QTL for the SAR. PHYB is the strongest candidate gene for SAR1 QTL. Here we studied the function of two polymorphic indels in the promoter region, a GGGR deletion, and three non-synonymous polymorphisms on the PHYB coding region compared with the Col-0 reference genome. To decipher the contribution and relevance of each PHYB-Pat polymorphism, we constructed transgenic lines with single or double polymorphisms by using Col-0 as a reference genome. We found that single polymorphisms in the coding region of PHYB have discrete functions in seed germination, seedling development, and shade avoidance response. These results suggest distinct functions for each PHYB polymorphism to the adjustment of plant development to variable light conditions.
Keywords: Arabidopsis; PHYB; QTL mapping; natural genetic variation; shade avoidance response.
Copyright © 2022 Ruiz-Diaz, Matsusaka, Cascales, Sánchez, Sánchez-Lamas, Cerdán and Botto.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time.Plant Physiol. 1999 Mar;119(3):909-15. doi: 10.1104/pp.119.3.909. Plant Physiol. 1999. PMID: 10069829 Free PMC article.
-
The receptor-like kinase ERECTA contributes to the shade-avoidance syndrome in a background-dependent manner.Ann Bot. 2013 May;111(5):811-9. doi: 10.1093/aob/mct038. Epub 2013 Feb 26. Ann Bot. 2013. PMID: 23444123 Free PMC article.
-
A single haplotype hyposensitive to light and requiring strong vernalization dominates Arabidopsis thaliana populations in Patagonia, Argentina.Mol Ecol. 2017 Jul;26(13):3389-3404. doi: 10.1111/mec.14107. Epub 2017 May 2. Mol Ecol. 2017. PMID: 28316114
-
Hormonal Regulation in Shade Avoidance.Front Plant Sci. 2017 Sep 4;8:1527. doi: 10.3389/fpls.2017.01527. eCollection 2017. Front Plant Sci. 2017. PMID: 28928761 Free PMC article. Review.
-
Seedling development in buckwheat and the discovery of the photomorphogenic shade-avoidance response.Plant Biol (Stuttg). 2013 Nov;15(6):931-40. doi: 10.1111/plb.12077. Epub 2013 Sep 24. Plant Biol (Stuttg). 2013. PMID: 24112603 Review.
References
-
- Arana M. V., Sánchez-Lamas M., Strasser B., Ibarra S. E., Cerdán P. D., Botto J. F., et al. . (2014). Functional diversity of phytochrome family in the control of light and gibberellin-mediated germination in Arabidopsis. Plant Cell Environ. 37, 2014–2023. doi: 10.1111/pce.12286, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

