Pathogenic genetic variants identified in Australian families with paediatric cataract
- PMID: 36161833
- PMCID: PMC9422809
- DOI: 10.1136/bmjophth-2022-001064
Pathogenic genetic variants identified in Australian families with paediatric cataract
Abstract
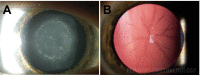
Objective: Paediatric (childhood or congenital) cataract is an opacification of the normally clear lens of the eye and has a genetic basis in at least 18% of cases in Australia. This study aimed to replicate clinical gene screening to identify variants likely to be causative of disease in an Australian patient cohort.
Methods and analysis: Sixty-three reported isolated cataract genes were screened for rare coding variants in 37 Australian families using genome sequencing.
Results: Disease-causing variants were confirmed in eight families with variant classification as 'likely pathogenic'. This included novel variants PITX3 p.(Ter303LeuextTer100), BFSP1 p.(Glu375GlyfsTer2), and GJA8 p.(Pro189Ser), as well as, previously described variants identified in genes GJA3, GJA8, CRYAA, BFSP1, PITX3, COL4A1 and HSF4. Additionally, eight variants of uncertain significance with evidence towards pathogenicity were identified in genes: GJA3, GJA8, LEMD2, PRX, CRYBB1, BFSP2, and MIP.
Conclusion: These findings expand the genotype-phenotype correlations of both pathogenic and benign variation in cataract-associated genes. They further emphasise the need to develop additional evidence such as functional assays and variant classification criteria specific to paediatric cataract genes to improve interpretation of variants and molecular diagnosis in patients.
Keywords: Child health (paediatrics); Experimental & laboratory; Genetics; Lens and zonules.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
